Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes
- PMID: 11717436
- PMCID: PMC61118
- DOI: 10.1073/pnas.241341898
Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes
Abstract
A new group of long terminal repeats (LTR) retrotransposons, termed terminal-repeat retrotransposons in miniature (TRIM), are described that are present in both monocotyledonous and dicotyledonous plant. TRIM elements have terminal direct repeat sequences between approximately 100 and 250 bp in length that encompass an internal domain of approximately 100-300 bp. The internal domain contains primer binding site and polypurine tract motifs but lacks the coding domains required for mobility. Thus TRIM elements are not capable of autonomous transposition and probably require the help of mobility-related proteins encoded by other retrotransposons. The structural organization of TRIM elements suggests an evolutionary relationship to either LTR retrotransposons or retroviruses. The past mobility of TRIM elements is indicated by the presence of flanking 5-bp direct repeats found typically at LTR retrotransposon insertion sites, the high degree of sequence conservation between elements from different genomic locations, and the identification of related to empty sites (RESites). TRIM elements seem to be involved actively in the restructuring of plant genomes, affecting the promoter, coding region and intron-exon structure of genes. In solanaceous species and maize, TRIM elements provided target sites for further retrotransposon insertions. In Arabidopsis, evidence is provided that the TRIM element also can be involved in the transduction of host genes.
Figures

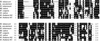
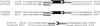


References
-
- Kunze R, Saedler H, Lonnig W E. Adv Bot Res. 1997;27:331–470.
-
- Boeke J D, Corces V G. Ann Rev Microbiol. 1989;43:403–434. - PubMed
-
- Boeke J D, Stoye J P. In: Retroviruses. Varmus H, Hughes S, Coffin J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 343–345. - PubMed
-
- Kumar A, Bennetzen J L. Annu Rev Genet. 1999;33:479–532. - PubMed
-
- Kidwell M G, Lisch D. Trends Ecol Evol. 2000;15:95–99. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

