Identification and characterization of a mitochondrial thioredoxin system in plants
- PMID: 11717467
- PMCID: PMC61182
- DOI: 10.1073/pnas.241340898
Identification and characterization of a mitochondrial thioredoxin system in plants
Abstract
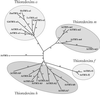
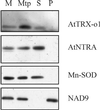
Plants possess two well described thioredoxin systems: a cytoplasmic system including several thioredoxins and an NADPH-dependent thioredoxin reductase and a specific chloroplastic system characterized by a ferredoxin-dependent thioredoxin reductase. On the basis of biochemical activities, plants also are supposed to have a mitochondrial thioredoxin system as described in yeast and mammals, but no gene encoding plant mitochondrial thioredoxin or thioredoxin reductase has been identified yet. We report the characterization of a plant thioredoxin system located in mitochondria. Arabidopsis thaliana genome sequencing has revealed numerous thioredoxin genes among which we have identified AtTRX-o1, a gene encoding a thioredoxin with a potential mitochondrial transit peptide. AtTRX-o1 and a second gene, AtTRX-o2, define, on the basis of the sequence and intron positions, a new thioredoxin type up to now specific to plants. We also have characterized AtNTRA, a gene encoding a protein highly similar to the previously described cytosolic NADPH-dependent thioredoxin reductase AtNTRB but with a putative presequence for import into mitochondria. Western blot analysis of A. thaliana subcellular and submitochondrial fractions and in vitro import experiments show that AtTRX-o1 and AtNTRA are targeted to the mitochondrial matrix through their cleavable N-terminal signal. The two proteins truncated to the estimated mature forms were produced in Escherichia coli; AtTRX-o1 efficiently reduces insulin in the presence of DTT and is reduced efficiently by AtNTRA and NADPH. Therefore, the thioredoxin and the NADPH-dependent thioredoxin reductase described here are proposed to constitute a functional plant mitochondrial thioredoxin system.
Figures




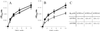
References
-
- Laurent T C, Moore E C, Reichard P. J Biol Chem. 1964;239:3436–3444. - PubMed
-
- Arner E S, Holmgren A. Eur J Biochem. 2000;267:6102–6109. - PubMed
-
- Meyer, Y., Vignols, F. & Reichheld, J.-P. (2001) Methods Enzymol., in press. - PubMed
-
- Pedrajas J R, Kosmidou E, Miranda-Vizuete A, Gustafsson J A, Wright A P, Spyrou G. J Biol Chem. 1999;274:6366–6373. - PubMed
-
- Spyrou G, Enmark E, Miranda-Vizuete A, Gustafsson J. J Biol Chem. 1997;272:2936–2941. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

