Percent G+C profiling accurately reveals diet-related differences in the gastrointestinal microbial community of broiler chickens
- PMID: 11722920
- PMCID: PMC93357
- DOI: 10.1128/AEM.67.12.5656-5667.2001
Percent G+C profiling accurately reveals diet-related differences in the gastrointestinal microbial community of broiler chickens
Abstract
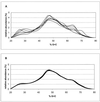
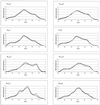
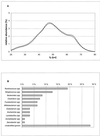
Broiler chickens from eight commercial farms in Southern Finland were analyzed for the structure of their gastrointestinal microbial community by a nonselective DNA-based method, percent G+C-based profiling. The bacteriological impact of the feed source and in-farm whole-wheat amendment of the diet was assessed by percent G+C profiling. Also, a phylogenetic 16S rRNA gene (rDNA)-based study was carried out to aid in interpretation of the percent G+C profiles. This survey showed that most of the 16S rDNA sequences found could not be assigned to any previously known bacterial genus or they represented an unknown species of one of the taxonomically heterogeneous genera, such as Ruminococcus or Clostridium. The data from bacterial community profiling were analyzed by t-test, multiple linear regression, and principal-component statistical approaches. The percent G+C profiling method with appropriate statistical analyses detected microbial community differences smaller than 10% within each 5% increment of the percent G+C profiles. Diet turned out to be the strongest determinant of the cecal bacterial community structure. Both the source of feed and local feed amendment changed the bacteriological profile significantly, whereas profiles of individual farms with identical feed regimens hardly differed from each other. This suggests that the management of typical Finnish farms is relatively uniform or that hygiene on the farm, in fact, has little impact on the structure of the cecal bacterial community. Therefore, feed compounders should have a significant role in the modulation of gut microflora and consequently in prevention of gastrointestinal disorders in farm animals.
Figures


References
-
- Afifi A A, Clark V. Principal component analysis. In: Afifi A A, Clark V, editors. Computer-aided multivariate analysis. New York, N.Y: Chapman & Hall; 1990. pp. 371–394.
-
- Apajalahti J H A, Särkilahti L K, Mäki B R E, Heikkinen J P, Nurminen P H, Holben W E. Effective recovery of bacterial DNA and percent-guanine-plus-cytosine-based analysis of community structure in the gastrointestinal tract of broiler chickens. Appl Environ Microbiol. 1998;64:4084–4088. - PMC - PubMed
-
- Brock T D, Madigan M T. Biology of microorganisms. Englewood Cliffs, N.J: Prentice Hall; 1991.
-
- Burr M D, Josephson K L, Pepper I L. An evaluation of ERIC PCR and AP PCR fingerprinting for discriminating Salmonella serotypes. Lett Appl Microbiol. 1998;27:24–30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

