In situ detection, isolation, and physiological properties of a thin filamentous microorganism abundant in methanogenic granular sludges: a novel isolate affiliated with a clone cluster, the green non-sulfur bacteria, subdivision I
- PMID: 11722931
- PMCID: PMC93368
- DOI: 10.1128/AEM.67.12.5740-5749.2001
In situ detection, isolation, and physiological properties of a thin filamentous microorganism abundant in methanogenic granular sludges: a novel isolate affiliated with a clone cluster, the green non-sulfur bacteria, subdivision I
Abstract
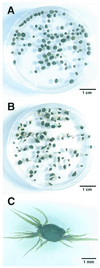
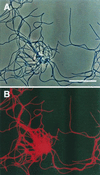
We previously showed that very thin filamentous bacteria affiliated with the division green non-sulfur bacteria were abundant in the outermost layer of thermophilic methanogenic sludge granules fed with sucrose and several low-molecular-weight fatty acids (Y. Sekiguchi, Y. Kamagata, K. Nakamura, A. Ohashi, H. Harada, Appl. Environ. Microbiol. 65:1280-1288, 1999). Further 16S ribosomal DNA (rDNA) cloning-based analysis revealed that the microbes were classified within a unique clade, green non-sulfur bacteria (GNSB) subdivision I, which contains a number of 16S rDNA clone sequences from various environmental samples but no cultured representatives. To investigate their function in the community and physiological traits, we attempted to isolate the yet-to-be-cultured microbes from the original granular sludge. The first attempt at isolation from the granules was, however, not successful. In the other thermophilic reactor that had been treating fried soybean curd-manufacturing wastewater, we found filamentous microorganisms to outgrow, resulting in the formation of projection-like structures on the surface of granules, making the granules look like sea urchins. 16S rDNA-cloning analysis combined with fluorescent in situ hybridization revealed that the projections were comprised of the uncultured filamentous cells affiliated with the GNSB subdivision I and Methanothermobacter-like cells and the very ends of the projections were comprised solely of the filamentous cells. By using the tip of the projection as the inoculum for primary enrichment, a thermophilic, strictly anaerobic, filamentous bacterium, designated strain UNI-1, was successfully isolated with a medium supplemented with sucrose and yeast extract. The strain was a very slow growing bacterium which is capable of utilizing only a limited range of carbohydrates in the presence of yeast extract and produced hydrogen from these substrates. The growth was found to be significantly stimulated when the strain was cocultured with a hydrogen-utilizing methanogen, Methanothermobacter thermautotrophicus, suggesting that the strain is a sugar-fermenting bacterium, the growth of which is dependent on hydrogen consumers in the granules.
Figures




References
-
- Alphenaar P A. Anaerobic granular sludge: characterization and factors affecting its functioning. Ph.D. thesis. Wageningen, The Netherlands: Wageningen Agricultural University; 1994.
-
- Chandler D P, Brockman F J, Bailey T J, Fredrickson J K. Phylogenetic diversity of Archaea and Bacteria in a deep subsurface pleosol. Microb Ecol. 1998;36:37–50. - PubMed
-
- Daims H, Breul A, Amann R, Schleifer K-H, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

