Cultivation-independent, semiautomatic determination of absolute bacterial cell numbers in environmental samples by fluorescence in situ hybridization
- PMID: 11722938
- PMCID: PMC93375
- DOI: 10.1128/AEM.67.12.5810-5818.2001
Cultivation-independent, semiautomatic determination of absolute bacterial cell numbers in environmental samples by fluorescence in situ hybridization
Abstract
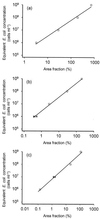
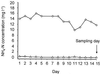
Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes has found widespread application for analyzing the composition of microbial communities in complex environmental samples. Although bacteria can quickly be detected by FISH, a reliable method to determine absolute numbers of FISH-stained cells in aggregates or biofilms has, to our knowledge, never been published. In this study we developed a semiautomated protocol to measure the concentration of bacteria (in cells per volume) in environmental samples by a combination of FISH, confocal laser scanning microscopy, and digital image analysis. The quantification is based on an internal standard, which is introduced by spiking the samples with known amounts of Escherichia coli cells. This method was initially tested with artificial mixtures of bacterial cultures and subsequently used to determine the concentration of ammonia-oxidizing bacteria in a municipal nitrifying activated sludge. The total number of ammonia oxidizers was found to be 9.8 x 10(7) +/- 1.9 x 10(7) cells ml(-1). Based on this value, the average in situ activity was calculated to be 2.3 fmol of ammonia converted to nitrite per ammonia oxidizer cell per h. This activity is within the previously determined range of activities measured with ammonia oxidizer pure cultures, demonstrating the utility of this quantification method for enumerating bacteria in samples in which cells are not homogeneously distributed.
Figures

References
-
- Amann R I. In situ identification of microorganisms by whole cell hybridization with rRNA-targeted nucleic acid probes. In: Akkeman A D C, van Elsas J D, de Bruigin F J, editors. Molecular microbial ecology manual. 3.3.6. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–15.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

