Synchronized activation and refolding of influenza hemagglutinin in multimeric fusion machines
- PMID: 11724823
- PMCID: PMC2150858
- DOI: 10.1083/jcb.200103005
Synchronized activation and refolding of influenza hemagglutinin in multimeric fusion machines
Abstract
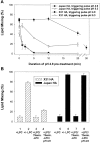
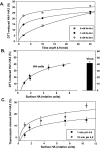
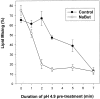
At the time of fusion, membranes are packed with fusogenic proteins. Do adjacent individual proteins interact with each other in the plane of the membrane? Or does each of these proteins serve as an independent fusion machine? Here we report that the low pH-triggered transition between the initial and final conformations of a prototype fusogenic protein, influenza hemagglutinin (HA), involves a preserved interaction between individual HAs. Although the HAs of subtypes H3 and H2 show notably different degrees of activation, for both, the percentage of low pH-activated HA increased with higher surface density of HA, indicating positive cooperativity. We propose that a concerted activation of HAs, together with the resultant synchronized release of their conformational energy, is an example of a general strategy of coordination in biological design, crucial for the functioning of multiprotein fusion machines.
Figures





Similar articles
-
Membrane fusion by surrogate receptor-bound influenza haemagglutinin.Virology. 1999 May 10;257(2):415-23. doi: 10.1006/viro.1999.9624. Virology. 1999. PMID: 10329552
-
Reversible stages of the low-pH-triggered conformational change in influenza virus hemagglutinin.EMBO J. 2002 Nov 1;21(21):5701-10. doi: 10.1093/emboj/cdf559. EMBO J. 2002. PMID: 12411488 Free PMC article.
-
Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin.Annu Rev Biochem. 2000;69:531-69. doi: 10.1146/annurev.biochem.69.1.531. Annu Rev Biochem. 2000. PMID: 10966468 Review.
-
Palmitoylation Contributes to Membrane Curvature in Influenza A Virus Assembly and Hemagglutinin-Mediated Membrane Fusion.J Virol. 2017 Oct 13;91(21):e00947-17. doi: 10.1128/JVI.00947-17. Print 2017 Nov 1. J Virol. 2017. PMID: 28794042 Free PMC article.
-
Acid-induced membrane fusion by the hemagglutinin protein and its role in influenza virus biology.Curr Top Microbiol Immunol. 2014;385:93-116. doi: 10.1007/82_2014_393. Curr Top Microbiol Immunol. 2014. PMID: 25007844 Free PMC article. Review.
Cited by
-
Influenza hemagglutinin drives viral entry via two sequential intramembrane mechanisms.Proc Natl Acad Sci U S A. 2020 Mar 31;117(13):7200-7207. doi: 10.1073/pnas.1914188117. Epub 2020 Mar 18. Proc Natl Acad Sci U S A. 2020. PMID: 32188780 Free PMC article.
-
Negative potentials across biological membranes promote fusion by class II and class III viral proteins.Mol Biol Cell. 2010 Jun 15;21(12):2001-12. doi: 10.1091/mbc.e09-10-0904. Epub 2010 Apr 28. Mol Biol Cell. 2010. PMID: 20427575 Free PMC article.
-
Measuring pKa of activation and pKi of inactivation for influenza hemagglutinin from kinetics of membrane fusion of virions and of HA expressing cells.Biophys J. 2002 Nov;83(5):2652-66. doi: 10.1016/S0006-3495(02)75275-2. Biophys J. 2002. PMID: 12414698 Free PMC article.
-
Virus membrane-fusion proteins: more than one way to make a hairpin.Nat Rev Microbiol. 2006 Jan;4(1):67-76. doi: 10.1038/nrmicro1326. Nat Rev Microbiol. 2006. PMID: 16357862 Free PMC article. Review.
-
Intracellular curvature-generating proteins in cell-to-cell fusion.Biochem J. 2011 Dec 1;440(2):185-93. doi: 10.1042/BJ20111243. Biochem J. 2011. PMID: 21895608 Free PMC article.
References
-
- Bron, R., A. Ortiz, J. Dijkstra, T. Stegmann, and J. Wilschut. 1993. Preparation, properties, and applications of reconstituted influenza virus envelopes (virosomes). Methods Enzymol. 220:313–331. - PubMed
-
- Bullough, P.A., F.M. Hughson, J.J. Skehel, and D.C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 371:37–43. - PubMed
-
- Carr, C.M., and P.S. Kim. 1993. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 73:823–832. - PubMed

