PCR-based detection and identification of Burkholderia cepacia complex pathogens in sputum from cystic fibrosis patients
- PMID: 11724828
- PMCID: PMC88532
- DOI: 10.1128/JCM.39.12.4247-4255.2001
PCR-based detection and identification of Burkholderia cepacia complex pathogens in sputum from cystic fibrosis patients
Abstract
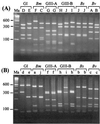
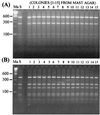
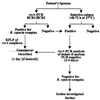
PCR amplification of the recA gene followed by restriction fragment length polymorphism (RFLP) analysis was investigated for the rapid detection and identification of Burkholderia cepacia complex genomovars directly from sputum. Successful amplification of the B. cepacia complex recA gene from cystic fibrosis (CF) patient sputum samples containing B. cepacia genomovar I, Burkholderia multivorans, B. cepacia genomovar III, Burkholderia stabilis, and Burkholderia vietnamiensis was demonstrated. In addition, the genomovar identifications determined directly from sputum were the same as those obtained after selective culturing. Sensitivity experiments revealed that recA-based PCR could reliably detect B. cepacia complex organisms to concentrations of 10(6) CFU g of sputum(-1). To fully assess the diagnostic value of the method, sputum samples from 100 CF patients were screened for B. cepacia complex infection by selective culturing and recA-based PCR. Selective culturing identified 19 samples with presumptive B. cepacia complex infection, which was corroborated by phenotypic analyses. Of the culture-positive sputum samples, 17 were also detected directly by recA-based PCR, while 2 samples were negative. The isolates cultured from both recA-negative sputum samples were subsequently identified as Burkholderia gladioli. RFLP analysis of the recA amplicons revealed 2 patients (12%) infected with B. multivorans, 11 patients (65%) infected with B. cepacia genomovar III-A, and 4 patients (23%) infected with B. cepacia genomovar III-B. These results demonstrate the potential of recA-based PCR-RFLP analysis for the rapid detection and identification of B. cepacia complex genomovars directly from sputum. Where the sensitivity of the assay proves a limitation, sputum samples can be analyzed by selective culturing followed by recA-based analysis of the isolate.
Figures

Similar articles
-
Molecular characterization of Burkholderia cepacia isolates from cystic fibrosis (CF) patients in an Italian CF center.Res Microbiol. 2003 Sep;154(7):491-8. doi: 10.1016/S0923-2508(03)00145-1. Res Microbiol. 2003. PMID: 14499935
-
DNA-Based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III.J Clin Microbiol. 2000 Sep;38(9):3165-73. doi: 10.1128/JCM.38.9.3165-3173.2000. J Clin Microbiol. 2000. PMID: 10970351 Free PMC article.
-
Phenotypic methods for determining genomovar status of the Burkholderia cepacia complex.J Clin Microbiol. 2001 Mar;39(3):1073-8. doi: 10.1128/JCM.39.3.1073-1078.2001. J Clin Microbiol. 2001. PMID: 11230429 Free PMC article.
-
[Burkholderia cepacia: dangers of a phytopathogen organism for patients with cystic fibrosis].Ann Biol Clin (Paris). 2001 May-Jun;59(3):259-69. Ann Biol Clin (Paris). 2001. PMID: 11397673 Review. French.
-
Burkholderia cepacia: medical, taxonomic and ecological issues.J Med Microbiol. 1996 Dec;45(6):395-407. doi: 10.1099/00222615-45-6-395. J Med Microbiol. 1996. PMID: 8958242 Review.
Cited by
-
Diversity of Burkholderia cepacia complex from the Moso bamboo (Phyllostachys edulis) rhizhosphere soil.Curr Microbiol. 2011 Feb;62(2):650-8. doi: 10.1007/s00284-010-9758-3. Epub 2010 Sep 30. Curr Microbiol. 2011. PMID: 20882285
-
Evaluation of different molecular and phenotypic methods for identification of environmental Burkholderia cepacia complex.World J Microbiol Biotechnol. 2019 Feb 9;35(3):39. doi: 10.1007/s11274-019-2614-0. World J Microbiol Biotechnol. 2019. PMID: 30739255
-
PCR-based detection of a cystic fibrosis epidemic strain of Pseudomonas Aeruginosa.Mol Diagn. 2003;7(3-4):195-200. doi: 10.1007/BF03260038. Mol Diagn. 2003. PMID: 15068391
-
Use of 16S rRNA gene sequencing for identification of nonfermenting gram-negative bacilli recovered from patients attending a single cystic fibrosis center.J Clin Microbiol. 2002 Oct;40(10):3793-7. doi: 10.1128/JCM.40.10.3793-3797.2002. J Clin Microbiol. 2002. PMID: 12354883 Free PMC article.
-
A practical molecular identification of nonfermenting Gram-negative bacteria from cystic fibrosis.Braz J Microbiol. 2018 Apr-Jun;49(2):422-428. doi: 10.1016/j.bjm.2017.07.002. Epub 2017 Nov 4. Braz J Microbiol. 2018. PMID: 29157900 Free PMC article.
References
-
- Campbell P, Phillips J A, Heidecker G J, Krishnamani M R S, Zahorchak R, Stull T L. Detection of Pseudomonas (Burkholderia) cepacia using PCR. Pediatr Pulmonol. 1995;20:44–49. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

