Okazaki fragment processing: modulation of the strand displacement activity of DNA polymerase delta by the concerted action of replication protein A, proliferating cell nuclear antigen, and flap endonuclease-1
- PMID: 11724925
- PMCID: PMC64676
- DOI: 10.1073/pnas.251193198
Okazaki fragment processing: modulation of the strand displacement activity of DNA polymerase delta by the concerted action of replication protein A, proliferating cell nuclear antigen, and flap endonuclease-1
Abstract
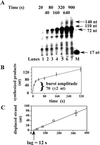
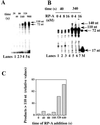
DNA polymerase (pol) delta is essential for both leading and lagging strand DNA synthesis during chromosomal replication in eukaryotes. Pol delta has been implicated in the Okazaki fragment maturation process for the extension of the newly synthesized fragment and for the displacement of the RNA/DNA segment of the preexisting downstream fragment generating an intermediate flap structure that is the target for the Dna2 and flap endonuclease-1 (Fen 1) endonucleases. Using a single-stranded minicircular template with an annealed RNA/DNA primer, we could measure strand displacement by pol delta coupled to DNA synthesis. Our results suggested that pol delta alone can displace up to 72 nucleotides while synthesizing through a double-stranded DNA region in a distributive manner. Proliferating cell nuclear antigen (PCNA) reduced the template dissociation rate of pol delta, thus increasing the processivity of both synthesis and strand displacement, whereas replication protein A (RP-A) limited the size of the displaced fragment down to 20-30 nucleotides, by generating a "locked" flap DNA structure, which was a substrate for processing of the displaced fragment by Fen 1 into a ligatable product. Our data support a model for Okazaki fragment processing where the strand displacement activity of DNA polymerase delta is modulated by the concerted action of PCNA, RP-A and Fen 1.
Figures



References
-
- Hindges R, Hübscher U. Biol Chem. 1997;378:345–362. - PubMed
-
- Waga S, Stillman B. Nature (London) 1994;369:207–212. - PubMed
-
- Waga S, Stillman B. Annu Rev Biochem. 1998;67:721–751. - PubMed
-
- Mossi R, Keller R C, Ferrari E, Hübscher U. J Mol Biol. 2000;295:803–814. - PubMed
-
- Maga G, Stucki M, Spadari S, Hübscher U. J Mol Biol. 2000;295:791–801. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

