Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF
- PMID: 11724935
- PMCID: PMC64679
- DOI: 10.1073/pnas.251421398
Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF
Abstract
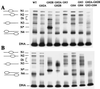
Nucleosome Remodeling Factor (NURF) is an ATP-dependent nucleosome remodeling complex that alters chromatin structure by catalyzing nucleosome sliding, thereby exposing DNA sequences previously associated with nucleosomes. We systematically studied how the unstructured N-terminal residues of core histones (the N-terminal histone tails) influence nucleosome sliding. We used bacterially expressed Drosophila histones to reconstitute hybrid nucleosomes lacking one or more histone N-terminal tails. Unexpectedly, we found that removal of the N-terminal tail of histone H2B promoted uncatalyzed nucleosome sliding during native gel electrophoresis. Uncatalyzed nucleosome mobility was enhanced by additional removal of other histone tails but was not affected by hyperacetylation of core histones by p300. In addition, we found that the N-terminal tail of the histone H4 is specifically required for ATP-dependent catalysis of nucleosome sliding by NURF. Alanine scanning mutagenesis demonstrated that H4 residues 16-KRHR-19 are critical for the induction of nucleosome mobility, revealing a histone tail motif that regulates NURF activity. An exchange of histone tails between H4 and H3 impaired NURF-induced sliding of the mutant nucleosome, indicating that the location of the KRHR motif in relation to global nucleosome structure is functionally important. Our results provide functions for the N-terminal histone tails in regulating the mobility of nucleosomes.
Figures

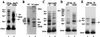


Similar articles
-
Role of histone tails in nucleosome remodeling by Drosophila NURF.EMBO J. 1997 Aug 1;16(15):4717-26. doi: 10.1093/emboj/16.15.4717. EMBO J. 1997. PMID: 9303316 Free PMC article.
-
p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone.Genes Dev. 2000 Aug 1;14(15):1899-907. Genes Dev. 2000. PMID: 10921904 Free PMC article.
-
Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI.Mol Cell Biol. 2001 Feb;21(3):875-83. doi: 10.1128/MCB.21.3.875-883.2001. Mol Cell Biol. 2001. PMID: 11154274 Free PMC article.
-
A modified epigenetics toolbox to study histone modifications on the nucleosome core.Chembiochem. 2011 Jan 24;12(2):308-13. doi: 10.1002/cbic.201000617. Epub 2010 Dec 29. Chembiochem. 2011. PMID: 21243718 Review.
-
Chromatin remodeling complexes: ATP-dependent machines in action.Biochem Cell Biol. 2005 Aug;83(4):405-17. doi: 10.1139/o05-115. Biochem Cell Biol. 2005. PMID: 16094444 Review.
Cited by
-
Quantitative determination of binding of ISWI to nucleosomes and DNA shows allosteric regulation of DNA binding by nucleotides.Biochemistry. 2014 Jul 15;53(27):4334-45. doi: 10.1021/bi500224t. Epub 2014 Jun 30. Biochemistry. 2014. PMID: 24898734 Free PMC article.
-
Concerted regulation of ISWI by an autoinhibitory domain and the H4 N-terminal tail.Elife. 2017 Jan 21;6:e21477. doi: 10.7554/eLife.21477. Elife. 2017. PMID: 28109157 Free PMC article.
-
The yeast ISW1b ATP-dependent chromatin remodeler is critical for nucleosome spacing and dinucleosome resolution.Sci Rep. 2021 Feb 18;11(1):4195. doi: 10.1038/s41598-021-82842-9. Sci Rep. 2021. PMID: 33602956 Free PMC article.
-
A cassette of N-terminal amino acids of histone H2B are required for efficient cell survival, DNA repair and Swi/Snf binding in UV irradiated yeast.Nucleic Acids Res. 2010 Mar;38(5):1450-60. doi: 10.1093/nar/gkp1074. Epub 2009 Dec 9. Nucleic Acids Res. 2010. PMID: 20007597 Free PMC article.
-
A nucleotide-driven switch regulates flanking DNA length sensing by a dimeric chromatin remodeler.Mol Cell. 2015 Mar 5;57(5):850-859. doi: 10.1016/j.molcel.2015.01.008. Epub 2015 Feb 12. Mol Cell. 2015. PMID: 25684208 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

