Helicobacter pylori genetic diversity within the gastric niche of a single human host
- PMID: 11724955
- PMCID: PMC64732
- DOI: 10.1073/pnas.251551698
Helicobacter pylori genetic diversity within the gastric niche of a single human host
Abstract
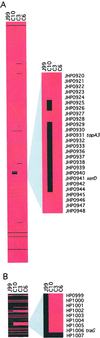
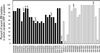
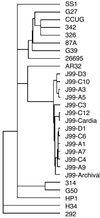
Isolates of the gastric pathogen Helicobacter pylori harvested from different individuals are highly polymorphic. Strain variation also has been observed within a single host. To more fully ascertain the extent of H. pylori genetic diversity within the ecological niche of its natural host, we harvested additional isolates of the sequenced H. pylori strain J99 from its human source patient after a 6-year interval. Randomly amplified polymorphic DNA PCR and DNA sequencing of four unlinked loci indicated that these isolates were closely related to the original strain. In contrast, microarray analysis revealed differences in genetic content among all of the isolates that were not detected by randomly amplified polymorphic DNA PCR or sequence analysis. Several ORFs from loci scattered throughout the chromosome in the archival strain did not hybridize with DNA from the recent strains, including multiple ORFs within the J99 plasticity zone. In addition, DNA from the recent isolates hybridized with probes for ORFs specific for the other fully sequenced H. pylori strain 26695, including a putative traG homolog. Among the additional J99 isolates, patterns of genetic diversity were distinct both when compared with each other and to the original prototype isolate. These results indicate that within an apparently homogeneous population, as determined by macroscale comparison and nucleotide sequence analysis, remarkable genetic differences exist among single-colony isolates of H. pylori. Direct evidence that H. pylori has the capacity to lose and possibly acquire exogenous DNA is consistent with a model of continuous microevolution within its cognate host.
Figures

References
-
- Blaser M J, Perez-Perez G I, Kleanthous H, Cover T L, Peek R M, Chyou P H, Stemmermann G N, Nomura A. Cancer Res. 1995;55:2111–2115. - PubMed
-
- Nomura A, Stemmermann G N, Chyou P H, Kato I, Pérez-Pérez G I, Blaser M J. N Engl J Med. 1991;325:1132–1136. - PubMed
-
- Parsonnet J, Friedman G D, Vandersteen D P, Chang Y, Vogelman J H, Orentreich N, Sibley R K. N Engl J Med. 1991;325:1127–1131. - PubMed
-
- Peterson W L. N Engl J Med. 1991;324:1043–1048. - PubMed
-
- Taylor D N, Blaser M J. Epidemiol Rev. 1991;13:42–59. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

