Enzyme-like proteins by computational design
- PMID: 11724958
- PMCID: PMC64672
- DOI: 10.1073/pnas.251555398
Enzyme-like proteins by computational design
Abstract
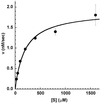
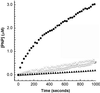
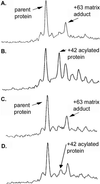
We report the development and initial experimental validation of a computational design procedure aimed at generating enzyme-like protein catalysts called "protozymes." Our design approach utilizes a "compute and build" strategy that is based on the physical/chemical principles governing protein stability and catalytic mechanism. By using the catalytically inert 108-residue Escherichia coli thioredoxin as a scaffold, the histidine-mediated nucleophilic hydrolysis of p-nitrophenyl acetate as a model reaction, and the ORBIT protein design software to compute sequences, an active site scan identified two promising catalytic positions and surrounding active-site mutations required for substrate binding. Experimentally, both candidate protozymes demonstrated catalytic activity significantly above background. One of the proteins, PZD2, displayed "burst" phase kinetics at high substrate concentrations, consistent with the formation of a stable enzyme intermediate. The kinetic parameters of PZD2 are comparable to early catalytic Abs. But, unlike catalytic Ab design, our design procedure is independent of fold, suggesting a possible mechanism for examining the relationships between protein fold and the evolvability of protein function.
Figures

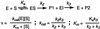

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

