A transcriptional roadmap to wood formation
- PMID: 11724959
- PMCID: PMC64750
- DOI: 10.1073/pnas.261293398
A transcriptional roadmap to wood formation
Abstract
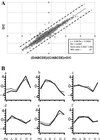
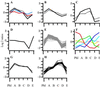
The large vascular meristem of poplar trees with its highly organized secondary xylem enables the boundaries between different developmental zones to be easily distinguished. This property of wood-forming tissues allowed us to determine a unique tissue-specific transcript profile for a well defined developmental gradient. RNA was prepared from different developmental stages of xylogenesis for DNA microarray analysis by using a hybrid aspen unigene set consisting of 2,995 expressed sequence tags. The analysis revealed that the genes encoding lignin and cellulose biosynthetic enzymes, as well as a number of transcription factors and other potential regulators of xylogenesis, are under strict developmental stage-specific transcriptional regulation.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

