Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure
- PMID: 11726495
- PMCID: PMC125751
- DOI: 10.1093/emboj/20.23.6591
Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure
Abstract
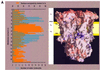
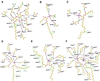
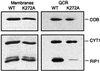
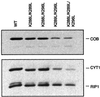
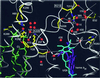
Biochemical data have shown that specific, tightly bound phospholipids are essential for activity of the cytochrome bc1 complex (QCR), an integral membrane protein of the respiratory chain. However, the structure and function of such phospholipids are not yet known. Here we describe five phospholipid molecules and one detergent molecule in the X-ray structure of yeast QCR at 2.3 A resolution. Their individual binding sites suggest specific roles in facilitating structural and functional integrity of the enzyme. Interestingly, a phosphatidylinositol molecule is bound in an unusual interhelical position near the flexible linker region of the Rieske iron-sulfur protein. Two possible proton uptake pathways at the ubiquinone reduction site have been identified: the E/R and the CL/K pathway. Remarkably, cardiolipin is positioned at the entrance to the latter. We propose that cardiolipin ensures structural integrity of the proton-conducting protein environment and takes part directly in proton uptake. Site-directed mutagenesis of ligating residues confirmed the importance of the phosphatidylinositol- and cardiolipin-binding sites.
Figures



References
-
- Berry E.A., Zhang,Z., Huang,L.S. and Kim,S.H. (1999) Structures of quinone-binding sites in bc complexes: functional implications. Biochem. Soc. Trans., 27, 565–572. - PubMed
-
- Berry E.A., Guergova-Kuras,M., Huang,L. and Crofts,A.R. (2000) Structure and function of cytochrome bc complexes. Annu. Rev. Biochem., 69, 1005–1075. - PubMed
-
- Beyer K. and Nuscher,B. (1996) Specific cardiolipin binding interferes with labeling of sulfhydryl residues in the adenosine diphosphate/adenosine triphosphate carrier protein from beef heart mitochondria. Biochemistry, 35, 15784–15790. - PubMed
-
- Brandt U. and Trumpower,B.L. (1994) The protonmotive Q cycle in mitochondria and bacteria. Crit. Rev. Biochem. Mol. Biol., 29, 165–197. - PubMed
-
- Brandt U., Uribe,S., Schägger,H. and Trumpower,B.L. (1994) Isolation and characterization of QCR10, the nuclear gene encoding the 8.5-kDa subunit 10 of the Saccharomyces cerevisiae cytochrome bc1 complex. J. Biol. Chem., 269, 12947–12953. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

