Yeast prion protein derivative defective in aggregate shearing and production of new 'seeds'
- PMID: 11726504
- PMCID: PMC125771
- DOI: 10.1093/emboj/20.23.6683
Yeast prion protein derivative defective in aggregate shearing and production of new 'seeds'
Abstract
According to the nucleated polymerization model, in vivo prion proliferation occurs via dissociation (shearing) of the huge prion polymers into smaller oligomeric 'seeds', initiating new rounds of prion replication. Here, we identify the deletion derivative of yeast prion protein Sup35 (Sup35-Delta22/69) that is specifically defective in aggregate shearing and 'seed' production. This derivative, [PSI+], previously thought to be unable to turn into a prion state, in fact retains the ability to form a prion ([PSI+](Delta22/69)) that can be maintained in selective conditions and transmitted by cytoplasmic infection (cytoduction), but which is mitotically unstable in non-selective conditions. MorePSI+](Delta22/69) retains its mitotic stability defect. The [PSI+](Delta22/69) cells contain more Sup35 protein in the insoluble fraction and form larger Sup35 aggregates compared with the conventional [PSI+] cells. Moderate excess of Hsp104 disaggregase increases transmission of the [PSI+](Delta22/69) prion, while excess Hsp70-Ssa chaperone antagonizes it, opposite to their effects on conventional [PSI+]. Our results shed light on the mechanisms determining the differences between transmissible prions and non-transmissible protein aggregates.
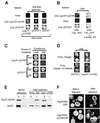
Figures

References
-
- Bonneaud N., Ozier-Kalogeropoulos,O., Li,G.Y., Labouesse,M., Minvielle-Sebastia,L. and Lacroute,F. (1991) A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E.coli shuttle vectors. Yeast, 7, 609–615. - PubMed
-
- Borchsenius A.S., Tchourikova,A.A. and Inge-Vechtomov,S.G. (2000) Recessive mutations in SUP35 and SUP45 genes coding for translation release factors affect chromosome stability in Saccharomyces cerevisiae. Curr. Genet., 37, 285–291. - PubMed
-
- Chernoff Y.O. (2001) Mutation processes at the protein level: is Lamarck back? Mutat. Res., 488, 39–64. - PubMed
-
- Chernoff Y.O., Derkach,I.L. and Inge-Vechtomov,S.G. (1993) Multicopy SUP35 gene induces de novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr. Genet., 24, 268–270. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

