Release of U18 snoRNA from its host intron requires interaction of Nop1p with the Rnt1p endonuclease
- PMID: 11726521
- PMCID: PMC125767
- DOI: 10.1093/emboj/20.23.6856
Release of U18 snoRNA from its host intron requires interaction of Nop1p with the Rnt1p endonuclease
Abstract
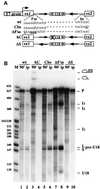
An external stem, essential for the release of small nucleolar RNAs (snoRNAs) from their pre-mRNAs, flanks the majority of yeast intron-encoded snoRNAs. Even if this stem is not a canonical Rnt1p substrate, several experiments have indicated that the Rnt1p endonuclease is required for snoRNA processing. To identify the factors necessary for processing of intron-encoded snoRNAs, we have raised in vitro extracts able to reproduce such activity. We found that snoRNP factors are associated with the snoRNA- coding region throughout all the processing steps, and that mutants unable to assemble snoRNPs have a processing-deficient phenotype. Specific depletion of Nop1p completely prevents U18 snoRNA synthesis, but does not affect processing of a dicistronic snoRNA-coding unit that has a canonical Rnt1p site. Correct cleavage of intron-encoded U18 and snR38 snoRNAs can be reproduced in vitro by incubating together purified Nop1p and Rnt1p. Pull-down experiments showed that the two proteins interact physically. These data indicate that cleavage of U18, snR38 and possibly other intron-encoded snoRNAs is a regulated process, since the stem is cleaved by the Rnt1p endonuclease only when snoRNP assembly has occurred.
Figures


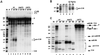


Similar articles
-
Identification of a novel element required for processing of intron-encoded box C/D small nucleolar RNAs in Saccharomyces cerevisiae.Mol Cell Biol. 2000 Feb;20(4):1311-20. doi: 10.1128/MCB.20.4.1311-1320.2000. Mol Cell Biol. 2000. PMID: 10648617 Free PMC article.
-
Genome-wide prediction and analysis of yeast RNase III-dependent snoRNA processing signals.Mol Cell Biol. 2005 Apr;25(8):2981-94. doi: 10.1128/MCB.25.8.2981-2994.2005. Mol Cell Biol. 2005. PMID: 15798187 Free PMC article.
-
The position of yeast snoRNA-coding regions within host introns is essential for their biosynthesis and for efficient splicing of the host pre-mRNA.RNA. 2007 Jan;13(1):138-50. doi: 10.1261/rna.251907. Epub 2006 Nov 29. RNA. 2007. PMID: 17135484 Free PMC article.
-
SnoRNP biogenesis meets Pre-mRNA splicing.Mol Cell. 2006 Sep 15;23(6):775-6. doi: 10.1016/j.molcel.2006.08.023. Mol Cell. 2006. PMID: 16973429 Review.
-
The vertebrate E1/U17 small nucleolar ribonucleoprotein particle.J Cell Biochem. 2006 Jun 1;98(3):486-95. doi: 10.1002/jcb.20821. J Cell Biochem. 2006. PMID: 16475166 Review.
Cited by
-
Processing of a dicistronic tRNA-snoRNA precursor: combined analysis in vitro and in vivo reveals alternate pathways and coupling to assembly of snoRNP.Plant Physiol. 2009 Jul;150(3):1598-610. doi: 10.1104/pp.109.137968. Epub 2009 May 6. Plant Physiol. 2009. PMID: 19420328 Free PMC article.
-
Mapping of in vivo cleavage sites uncovers a major role for yeast RNase III in regulating protein-coding genes.bioRxiv [Preprint]. 2025 Aug 13:2025.03.07.642061. doi: 10.1101/2025.03.07.642061. bioRxiv. 2025. PMID: 40832349 Free PMC article. Preprint.
-
The RNA catabolic enzymes Rex4p, Rnt1p, and Dbr1p show genetic interaction with trans-acting factors involved in processing of ITS1 in Saccharomyces cerevisiae pre-rRNA.RNA. 2004 Dec;10(12):1946-56. doi: 10.1261/rna.7155904. Epub 2004 Nov 3. RNA. 2004. PMID: 15525710 Free PMC article.
-
Loss of the Yeast SR Protein Npl3 Alters Gene Expression Due to Transcription Readthrough.PLoS Genet. 2015 Dec 22;11(12):e1005735. doi: 10.1371/journal.pgen.1005735. eCollection 2015 Dec. PLoS Genet. 2015. PMID: 26694144 Free PMC article.
-
A physical interaction between Gar1p and Rnt1pi is required for the nuclear import of H/ACA small nucleolar RNA-associated proteins.Mol Cell Biol. 2002 Jul;22(13):4792-802. doi: 10.1128/MCB.22.13.4792-4802.2002. Mol Cell Biol. 2002. PMID: 12052886 Free PMC article.
References
-
- AbouElela S.A., Igel,H. and Ares,M.,Jr (1996) RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell, 85, 115–124. - PubMed
-
- Bachellerie J.-P., Michot,B., Nicoloso,M., Balakin,A., Ni,J. and Fournier,M.J. (1995) Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem. Sci., 20, 261–264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

