Cell-specific proteins regulate viral RNA translation and virus-induced disease
- PMID: 11726525
- PMCID: PMC125770
- DOI: 10.1093/emboj/20.23.6899
Cell-specific proteins regulate viral RNA translation and virus-induced disease
Abstract
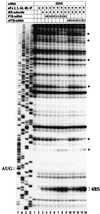
Translation initiation of the picornavirus genome is regulated by an internal ribosome entry site (IRES). The IRES of a neurovirulent picornavirus, the GDVII strain of Theiler's murine encephalomyelitis virus, requires polypyrimidine tract-binding protein (PTB) for its function. Although neural cells are deficient in PTB, they express a neural-specific homologue of PTB (nPTB). We now show that nPTB and PTB bind similarly to multiple sites in the GDVII IRES, rendering it competent for efficient translation initiation. Mutation of a PTB or nPTB site results in a more prominent decrease in nPTB than PTB binding, a decrease in activity of nPTB compared with PTB in promoting translation initiation, and attenuation of the neurovirulence of the virus without a marked effect on virus growth in non-neural cells. The addition of a second-site mutation in the mutant IRES generates a new PTB (nPTB) binding site, and restores nPTB binding, translation initiation and neurovirulence. We conclude that the tissue-specific expression and differential RNA-binding properties of PTB and nPTB are important determinants of cell-specific translational control and viral neurovirulence.
Figures


Similar articles
-
Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus.J Virol. 1997 Nov;71(11):8330-9. doi: 10.1128/JVI.71.11.8330-8339.1997. J Virol. 1997. PMID: 9343186 Free PMC article.
-
Polypyrimidine tract-binding protein inhibits translation of bip mRNA.J Mol Biol. 2000 Nov 24;304(2):119-33. doi: 10.1006/jmbi.2000.4179. J Mol Biol. 2000. PMID: 11080450
-
The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr.Mol Cell. 2003 Mar;11(3):757-71. doi: 10.1016/s1097-2765(03)00093-5. Mol Cell. 2003. PMID: 12667457
-
Mechanism of translation initiation on hepatitis C virus RNA.Princess Takamatsu Symp. 1995;25:111-9. Princess Takamatsu Symp. 1995. PMID: 8875615 Review.
-
Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein.Biochem Soc Trans. 2008 Aug;36(Pt 4):641-7. doi: 10.1042/BST0360641. Biochem Soc Trans. 2008. PMID: 18631133 Review.
Cited by
-
Picornaviruses and RNA Metabolism: Local and Global Effects of Infection.J Virol. 2019 Oct 15;93(21):e02088-17. doi: 10.1128/JVI.02088-17. Print 2019 Nov 1. J Virol. 2019. PMID: 31413128 Free PMC article. Review.
-
Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1.Mol Cell. 2007 Aug 3;27(3):420-34. doi: 10.1016/j.molcel.2007.06.016. Mol Cell. 2007. PMID: 17679092 Free PMC article.
-
Viral internal ribosome entry site elements: novel ribosome-RNA complexes and roles in viral pathogenesis.J Virol. 2003 Mar;77(5):2801-6. doi: 10.1128/jvi.77.5.2801-2806.2003. J Virol. 2003. PMID: 12584303 Free PMC article. Review. No abstract available.
-
RNA-binding proteins impacting on internal initiation of translation.Int J Mol Sci. 2013 Nov 1;14(11):21705-26. doi: 10.3390/ijms141121705. Int J Mol Sci. 2013. PMID: 24189219 Free PMC article. Review.
-
A single nucleotide in stem loop II of 5'-untranslated region contributes to virulence of enterovirus 71 in mice.PLoS One. 2011;6(11):e27082. doi: 10.1371/journal.pone.0027082. Epub 2011 Nov 1. PLoS One. 2011. PMID: 22069490 Free PMC article.
References
-
- Blyn L.B., Swiderek,K.M., Richards,O., Stahl,D.C., Semler,B.L. and Ehrenfeld,E. (1996) Poly(rC) binding protein 2 binds to stem–loop IV of the poliovirus RNA 5′ non-coding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl Acad. Sci. USA, 93, 11115–11120. - PMC - PubMed
-
- Dredge B.K., Polydorides,A.D. and Darnell,R.B. (2001) The splice of life: alternative splicing and neurological disease. Nature Rev. Neurosci., 2, 43–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

