Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex
- PMID: 11731475
- PMCID: PMC312842
- DOI: 10.1101/gad.926401
Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex
Abstract
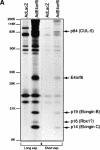
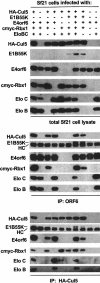
Although MDM2 plays a major role in regulating the stability of the p53 tumor suppressor protein, other poorly understood MDM2-independent pathways also exist. Human adenoviruses have evolved strategies to regulate p53 function and stability to permit efficient viral replication. One mechanism involves adenovirus E1B55K and E4orf6 proteins, which collaborate to target p53 for degradation. To determine the mechanism of this process, a multiprotein E4orf6-associated complex was purified and shown to contain a novel Cullin-containing E3 ubiquitin ligase that is (1) composed of Cullin family member Cul5, Elongins B and C, and the RING-H2 finger protein Rbx1(ROC1); (2) remarkably similar to the von Hippel-Lindau tumor suppressor and SCF (Skp1-Cul1/Cdc53-F-box) E3 ubiquitin ligase complexes; and (3) capable of stimulating ubiquitination of p53 in vitro in the presence of E1/E2 ubiquitin-activating and -conjugating enzymes. Cullins are activated by NEDD8 modification; therefore, to determine whether Cullin complexes are required for adenovirus-induced p53 degradation, studies were conducted in ts41 Chinese hamster ovary cells that are temperature sensitive for the NEDD8 pathway. E4orf6/E1B55K failed to induce the degradation of p53 at the nonpermissive temperature. Thus, our results identify a novel role for the Cullin-based machinery in regulation of p53.
Figures






Similar articles
-
Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53.Mol Cell Biol. 2004 Nov;24(21):9619-29. doi: 10.1128/MCB.24.21.9619-9629.2004. Mol Cell Biol. 2004. PMID: 15485928 Free PMC article.
-
Identification of integrin alpha3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex.J Virol. 2009 Jun;83(11):5329-38. doi: 10.1128/JVI.00089-09. Epub 2009 Mar 18. J Virol. 2009. PMID: 19297475 Free PMC article.
-
The adenovirus E4orf6 E3 ubiquitin ligase complex assembles in a novel fashion.Virology. 2007 Jul 20;364(1):36-44. doi: 10.1016/j.virol.2007.02.012. Epub 2007 Mar 23. Virology. 2007. PMID: 17367836
-
F-box proteins and protein degradation: an emerging theme in cellular regulation.Plant Mol Biol. 2000 Sep;44(2):123-8. doi: 10.1023/a:1006413007456. Plant Mol Biol. 2000. PMID: 11117256 Review.
-
The structure and regulation of Cullin 2 based E3 ubiquitin ligases and their biological functions.Cell Div. 2016 May 23;11:7. doi: 10.1186/s13008-016-0020-7. eCollection 2016. Cell Div. 2016. PMID: 27222660 Free PMC article. Review.
Cited by
-
En Guard! The Interactions between Adenoviruses and the DNA Damage Response.Viruses. 2020 Sep 7;12(9):996. doi: 10.3390/v12090996. Viruses. 2020. PMID: 32906746 Free PMC article. Review.
-
Integrative transcriptome analysis of SARS-CoV-2 human-infected cells combined with deep learning algorithms identifies two potential cellular targets for the treatment of coronavirus disease.Braz J Microbiol. 2023 Mar;54(1):53-68. doi: 10.1007/s42770-022-00875-2. Epub 2022 Nov 26. Braz J Microbiol. 2023. PMID: 36435956 Free PMC article.
-
A ubiquitin-specific protease possesses a decisive role for adenovirus replication and oncogene-mediated transformation.PLoS Pathog. 2013 Mar;9(3):e1003273. doi: 10.1371/journal.ppat.1003273. Epub 2013 Mar 28. PLoS Pathog. 2013. PMID: 23555268 Free PMC article.
-
The Human Adenovirus Type 5 E4orf6/E1B55K E3 Ubiquitin Ligase Complex Can Mimic E1A Effects on E2F.mSphere. 2015 Nov 11;1(1):e00014-15. doi: 10.1128/mSphere.00014-15. eCollection 2016 Jan-Feb. mSphere. 2015. PMID: 27303679 Free PMC article.
-
Ubiquitination at the interface of tumor viruses and DNA damage responses.Curr Opin Virol. 2018 Oct;32:40-47. doi: 10.1016/j.coviro.2018.08.017. Epub 2018 Sep 24. Curr Opin Virol. 2018. PMID: 30261451 Free PMC article. Review.
References
-
- Bacchetti S, Graham FL. Inhibition of cell proliferation by an adenovirus vector expressing the human wild type p53 protein. Int J Oncol. 1993;3:781–788. - PubMed
-
- Bargonetti J, Reynisdottir I, Friedman PN, Prives C. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes & Dev. 1992;6:1886–1898. - PubMed
-
- Bartek J, Lukas J. p27 destruction: Cks1 pulls the trigger. Nat Cell Biol. 2001;3:E95–E98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
