The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor
- PMID: 11731804
- PMCID: PMC2792006
- DOI: 10.1038/nsb737
The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor
Abstract
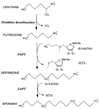
Polyamines are essential in all branches of life. Spermidine synthase (putrescine aminopropyltransferase, PAPT) catalyzes the biosynthesis of spermidine, a ubiquitous polyamine. The crystal structure of the PAPT from Thermotoga maritima (TmPAPT) has been solved to 1.5 A resolution in the presence and absence of AdoDATO (S-adenosyl-1,8-diamino-3-thiooctane), a compound containing both substrate and product moieties. This, the first structure of an aminopropyltransferase, reveals deep cavities for binding substrate and cofactor, and a loop that envelops the active site. The AdoDATO binding site is lined with residues conserved in PAPT enzymes from bacteria to humans, suggesting a universal catalytic mechanism. Other conserved residues act sterically to provide a structural basis for polyamine specificity. The enzyme is tetrameric; each monomer consists of a C-terminal domain with a Rossmann-like fold and an N-terminal beta-stranded domain. The tetramer is assembled using a novel barrel-type oligomerization motif.
Figures
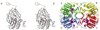


Similar articles
-
Crystal structure of Plasmodium falciparum spermidine synthase in complex with the substrate decarboxylated S-adenosylmethionine and the potent inhibitors 4MCHA and AdoDATO.J Mol Biol. 2007 Oct 12;373(1):167-77. doi: 10.1016/j.jmb.2007.07.053. Epub 2007 Aug 2. J Mol Biol. 2007. PMID: 17822713
-
Structure and mechanism of spermidine synthases.Biochemistry. 2007 Jul 17;46(28):8331-9. doi: 10.1021/bi602498k. Epub 2007 Jun 22. Biochemistry. 2007. PMID: 17585781
-
Binding and inhibition of human spermidine synthase by decarboxylated S-adenosylhomocysteine.Protein Sci. 2011 Nov;20(11):1836-44. doi: 10.1002/pro.717. Epub 2011 Sep 15. Protein Sci. 2011. PMID: 21898642 Free PMC article.
-
Aminopropyltransferases: function, structure and genetics.J Biochem. 2006 Jan;139(1):1-9. doi: 10.1093/jb/mvj019. J Biochem. 2006. PMID: 16428313 Review.
-
Use of aminopropyltransferase inhibitors and of non-metabolizable analogs to study polyamine regulation and function.Int J Biochem Cell Biol. 1995 May;27(5):425-42. doi: 10.1016/1357-2725(95)00007-c. Int J Biochem Cell Biol. 1995. PMID: 7641073 Review.
Cited by
-
Evolution of the key alkaloid enzyme putrescine N-methyltransferase from spermidine synthase.Front Plant Sci. 2013 Jul 29;4:260. doi: 10.3389/fpls.2013.00260. eCollection 2013. Front Plant Sci. 2013. PMID: 23908659 Free PMC article.
-
Crystal complexes of a predicted S-adenosylmethionine-dependent methyltransferase reveal a typical AdoMet binding domain and a substrate recognition domain.Protein Sci. 2003 Jul;12(7):1432-42. doi: 10.1110/ps.0302403. Protein Sci. 2003. PMID: 12824489 Free PMC article.
-
A polyamine metabolon involving aminopropyl transferase complexes in Arabidopsis.Plant Cell. 2002 Oct;14(10):2539-51. doi: 10.1105/tpc.004077. Plant Cell. 2002. PMID: 12368503 Free PMC article.
-
Recombinant RquA catalyzes the in vivo conversion of ubiquinone to rhodoquinone in Escherichia coli and Saccharomyces cerevisiae.Biochim Biophys Acta Mol Cell Biol Lipids. 2019 Sep;1864(9):1226-1234. doi: 10.1016/j.bbalip.2019.05.007. Epub 2019 May 21. Biochim Biophys Acta Mol Cell Biol Lipids. 2019. PMID: 31121262 Free PMC article.
-
Metagenomic Insights into the Regulatory Effects of Microbial Community on the Formation of Biogenic Amines and Volatile Flavor Components during the Brewing of Hongqu Rice Wine.Foods. 2023 Aug 16;12(16):3075. doi: 10.3390/foods12163075. Foods. 2023. PMID: 37628073 Free PMC article.
References
-
- Pegg AE, Poulin R, Coward JK. Int. J. Biochem. Cell Biol. 1995;27:425–442. - PubMed
-
- Lakanen JR, Pegg AE, Coward JK. J. Med. Chem. 1995;38:2714–2727. - PubMed
-
- Tang KC, Pegg AE, Coward JK. Biochem. Biophys. Res. Commun. 1980;96:1371–1377. - PubMed
-
- Rossmann MG, Liljas A, Branden C-I, Banaszak LK. In: Oxidation-reduction. Boyer PD, editor. New York: Academic Press; 1975. pp. 61–102.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
