Human-specific duplication and mosaic transcripts: the recent paralogous structure of chromosome 22
- PMID: 11731936
- PMCID: PMC419985
- DOI: 10.1086/338458
Human-specific duplication and mosaic transcripts: the recent paralogous structure of chromosome 22
Abstract
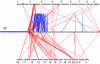
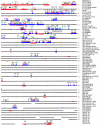
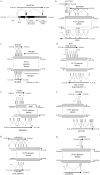
In recent decades, comparative chromosomal banding, chromosome painting, and gene-order studies have shown strong conservation of gross chromosome structure and gene order in mammals. However, findings from the human genome sequence suggest an unprecedented degree of recent (<35 million years ago) segmental duplication. This dynamism of segmental duplications has important implications in disease and evolution. Here we present a chromosome-wide view of the structure and evolution of the most highly homologous duplications (> or = 1 kb and > or = 90%) on chromosome 22. Overall, 10.8% (3.7/33.8 Mb) of chromosome 22 is duplicated, with an average sequence identity of 95.4%. To organize the duplications into tractable units, intron-exon structure and well-defined duplication boundaries were used to define 78 duplicated modules (minimally shared evolutionary segments) with 157 copies on chromosome 22. Analysis of these modules provides evidence for the creation or modification of 11 novel transcripts. Comparative FISH analyses of human, chimpanzee, gorilla, orangutan, and macaque reveal qualitative and quantitative differences in the distribution of these duplications--consistent with their recent origin. Several duplications appear to be human specific, including a approximately 400-kb duplication (99.4%-99.8% sequence identity) that transposed from chromosome 14 to the most proximal pericentromeric region of chromosome 22. Experimental and in silico data further support a pericentromeric gradient of duplications where the most recent duplications transpose adjacent to the centromere. Taken together, these data suggest that segmental duplications have been an ongoing process of primate genome evolution, contributing to recent gene innovation and the dynamic transformation of genome architecture within and among closely related species.
Figures
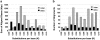



References
Electronic-Database Information
-
- RefSeq Database, http://www.ncbi.nlm.nih.gov/LocusLink/refseq.html (for accessions of the format NM_#####)
-
- RepeatMasker, http://repeatmasker.genome.washington.edu/
-
- Rocchi Lab Web site, http://www.biologia.uniba.it/22-paper/ (for all FISH images)
References
-
- Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, Cassidy SB, Driscoll DJ, Rogan PK, Schwartz S, Nicholls RD (1999) Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet 65:370–386 - PMC - PubMed
-
- Chen K, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault A, Lee C, Lupski J (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163 - PubMed
-
- Dorschner MO, Sybert VP, Weaver M, Pletcher BA, Stephens K (2000) NF1 microdeletion breakpoints are clustered at flanking repetitive sequences. Hum Mol Genet 9:35–46 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

