Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia
- PMID: 11733578
- PMCID: PMC2193523
- DOI: 10.1084/jem.194.11.1639
Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia
Abstract
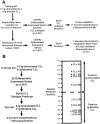
The most common human leukemia is B cell chronic lymphocytic leukemia (CLL), a malignancy of mature B cells with a characteristic clinical presentation but a variable clinical course. The rearranged immunoglobulin (Ig) genes of CLL cells may be either germ-line in sequence or somatically mutated. Lack of Ig mutations defined a distinctly worse prognostic group of CLL patients raising the possibility that CLL comprises two distinct diseases. Using genomic-scale gene expression profiling, we show that CLL is characterized by a common gene expression "signature," irrespective of Ig mutational status, suggesting that CLL cases share a common mechanism of transformation and/or cell of origin. Nonetheless, the expression of hundreds of other genes correlated with the Ig mutational status, including many genes that are modulated in expression during mitogenic B cell receptor signaling. These genes were used to build a CLL subtype predictor that may help in the clinical classification of patients with this disease.
Figures




References
-
- Hamblin, T.J., Z. Davis, A. Gardiner, D.G. Oscier, and F.K. Stevenson. 1999. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 94:1848–1854. - PubMed
-
- Damle, R.N., T. Wasil, F. Fais, F. Ghiotto, A. Valetto, S.L. Allen, A. Buchbinder, D. Budman, K. Dittmar, J. Kolitz, et al. 1999. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 94:1840–1847. - PubMed
-
- Jacob, J., G. Kelsoe, K. Rajewsky, and U. Weiss. 1991. Intraclonal generation of antibody mutants in germinal centers. Nature. 354:389–392. - PubMed
-
- Klein, U., T. Goossens, M. Fischer, H. Kanzler, A. Braeuninger, K. Rajewsky, and R. Kuppers. 1998. Somatic hypermutation in normal and transformed human B cells. Immunol. Rev. 162:261–281. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

