Three retrotransposon families in the genome of Giardia lamblia: two telomeric, one dead
- PMID: 11734649
- PMCID: PMC64710
- DOI: 10.1073/pnas.231494798
Three retrotransposon families in the genome of Giardia lamblia: two telomeric, one dead
Abstract
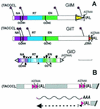
Transposable elements inhabiting eukaryotic genomes are generally regarded either as selfish DNA, which is selectively neutral to the host organism, or as parasitic DNA, deleterious to the host. Thus far, the only agreed-upon example of beneficial eukaryotic transposons is provided by Drosophila telomere-associated retrotransposons, which transpose directly to the chromosome ends and thereby protect them from degradation. This article reports the transposon content of the genome of the protozoan Giardia lamblia, one of the earliest-branching eukaryotes. A total of three non-long terminal repeat retrotransposon families have been identified, two of which are located at the ends of chromosomes, and the third one contains exclusively dead copies with multiple internal deletions, nucleotide substitutions, and frame shifts. No other reverse transcriptase- or transposase-related sequences were found. Thus, the entire genome of this protozoan, which is not known to reproduce sexually, contains only retrotransposons that are either confined to telomeric regions and possibly beneficial, or inactivated and completely nonfunctional.
Figures


Comment in
-
Another protozoan contributes to understanding telomeres and transposable elements.Proc Natl Acad Sci U S A. 2001 Dec 4;98(25):14195-7. doi: 10.1073/pnas.261567398. Proc Natl Acad Sci U S A. 2001. PMID: 11734638 Free PMC article. Review. No abstract available.
References
-
- Arkhipova I, Lyubomirskaya N, Ilyin Y. Drosophila Retrotransposons. Austin, TX: Landes; 1995.
-
- Capy P, Bazin C, Higuet D, Langin T. Dynamics and Evolution of Transposable Elements. Austin, TX: Landes; 1998.
-
- Mark Welch D, Meselson M. Science. 2000;288:1211–1215. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

