The N domain of Smad7 is essential for specific inhibition of transforming growth factor-beta signaling
- PMID: 11739411
- PMCID: PMC2150897
- DOI: 10.1083/jcb.200106023
The N domain of Smad7 is essential for specific inhibition of transforming growth factor-beta signaling
Abstract
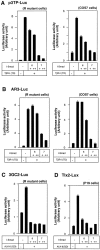
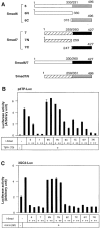
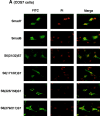
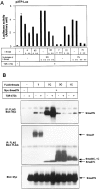
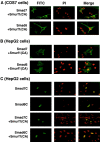
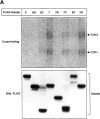
Inhibitory Smads (I-Smads) repress signaling by cytokines of the transforming growth factor-beta (TGF-beta) superfamily. I-Smads have conserved carboxy-terminal Mad homology 2 (MH2) domains, whereas the amino acid sequences of their amino-terminal regions (N domains) are highly divergent from those of other Smads. Of the two different I-Smads in mammals, Smad7 inhibited signaling by both TGF-beta and bone morphogenetic proteins (BMPs), whereas Smad6 was less effective in inhibiting TGF-beta signaling. Analyses using deletion mutants and chimeras of Smad6 and Smad7 revealed that the MH2 domains were responsible for the inhibition of both TGF-beta and BMP signaling by I-Smads, but the isolated MH2 domains of Smad6 and Smad7 were less potent than the full-length Smad7 in inhibiting TGF-beta signaling. The N domains of I-Smads determined the subcellular localization of these molecules. Chimeras containing the N domain of Smad7 interacted with the TGF-beta type I receptor (TbetaR-I) more efficiently, and were more potent in repressing TGF-beta signaling, than those containing the N domain of Smad6. The isolated N domain of Smad7 physically interacted with the MH2 domain of Smad7, and enhanced the inhibitory activity of the latter through facilitating interaction with TGF-beta receptors. The N domain of Smad7 thus plays an important role in the specific inhibition of TGF-beta signaling.
Figures








Similar articles
-
Roles for the MH2 domain of Smad7 in the specific inhibition of transforming growth factor-beta superfamily signaling.J Biol Chem. 2004 Jul 23;279(30):31568-74. doi: 10.1074/jbc.M313977200. Epub 2004 May 17. J Biol Chem. 2004. PMID: 15148321
-
NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) negatively regulates TGF-beta (transforming growth factor-beta) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-beta type I receptor.Biochem J. 2005 Mar 15;386(Pt 3):461-70. doi: 10.1042/BJ20040738. Biochem J. 2005. PMID: 15496141 Free PMC article.
-
Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation.J Biol Chem. 2001 Apr 20;276(16):12477-80. doi: 10.1074/jbc.C100008200. Epub 2001 Feb 13. J Biol Chem. 2001. PMID: 11278251
-
Regulation of transforming growth factor-beta signaling.Mol Cell Biol Res Commun. 2001 Nov;4(6):321-30. doi: 10.1006/mcbr.2001.0301. Mol Cell Biol Res Commun. 2001. PMID: 11703090 Review.
-
Two major Smad pathways in TGF-beta superfamily signalling.Genes Cells. 2002 Dec;7(12):1191-204. doi: 10.1046/j.1365-2443.2002.00599.x. Genes Cells. 2002. PMID: 12485160 Review.
Cited by
-
MyD88 Regulates the Expression of SMAD4 and the Iron Regulatory Hormone Hepcidin.Front Cell Dev Biol. 2018 Aug 31;6:105. doi: 10.3389/fcell.2018.00105. eCollection 2018. Front Cell Dev Biol. 2018. PMID: 30234111 Free PMC article.
-
Interaction between Smad7 and beta-catenin: importance for transforming growth factor beta-induced apoptosis.Mol Cell Biol. 2005 Feb;25(4):1475-88. doi: 10.1128/MCB.25.4.1475-1488.2005. Mol Cell Biol. 2005. PMID: 15684397 Free PMC article.
-
Smad7 regulates terminal maturation of chondrocytes in the growth plate.Dev Biol. 2013 Oct 15;382(2):375-84. doi: 10.1016/j.ydbio.2013.08.021. Epub 2013 Aug 29. Dev Biol. 2013. PMID: 23994637 Free PMC article.
-
Evolutionary roots of arginase expression and regulation.Front Immunol. 2014 Nov 7;5:544. doi: 10.3389/fimmu.2014.00544. eCollection 2014. Front Immunol. 2014. PMID: 25426114 Free PMC article. Review.
-
Post-Transcriptional Regulatory Crosstalk between MicroRNAs and Canonical TGF-β/BMP Signalling Cascades on Osteoblast Lineage: A Comprehensive Review.Int J Mol Sci. 2023 Mar 29;24(7):6423. doi: 10.3390/ijms24076423. Int J Mol Sci. 2023. PMID: 37047394 Free PMC article. Review.
References
-
- Bai, S., X. Shi, X. Yang, and X. Cao. 2000. Smad6 as a transcriptional corepressor. J. Biol. Chem. 275:8267–8270. - PubMed
-
- Ebisawa, T., M. Fukuchi, G. Murakami, T. Chiba, K. Tanaka, T. Imamura, and K. Miyazono. 2001. Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 276:12477–12480. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

