The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner
- PMID: 11739676
- PMCID: PMC135729
- DOI: 10.1128/jvi.76.1.105-117.2002
The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner
Abstract
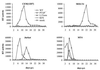
The p6 domain of human immunodeficiency virus type 1 (HIV-1) is located at the C terminus of the Gag precursor protein Pr55(Gag). Previous studies indicated that p6 plays a critical role in HIV-1 particle budding from virus-expressing HeLa cells. In this study, we performed a detailed mutational analysis of the N terminus of p6 to map the sequences required for efficient virus release. We observed that the highly conserved P-T/S-A-P motif located near the N terminus of p6 is remarkably sensitive to change; even conservative mutations in this sequence imposed profound virus release defects in HeLa cells. In contrast, single and double amino acid substitutions outside the P-T/S-A-P motif had no significant effect on particle release. The introduction of stop codons one or two residues beyond the P-T/S-A-P motif markedly impaired virion release, whereas truncation four residues beyond P-T/S-A-P had no effect on particle production in HeLa cells. By examining the effects of p6 mutation in biological and biochemical analyses and by electron microscopy, we defined the role of p6 in particle release and virus replication in a panel of T-cell and adherent cell lines and in primary lymphocytes and monocyte-derived macrophages. We demonstrated that the effects of p6 mutation on virus replication are markedly cell type dependent. Intriguingly, even in T-cell lines and primary lymphocytes in which p6 mutations block virus replication, these changes had little or no effect on particle release. However, p6-mutant particles produced in T-cell lines and primary lymphocytes exhibited a defect in virion-virion detachment, resulting in the production of tethered chains of virions. Virus release in monocyte-derived macrophages was markedly inhibited by p6 mutation. To examine further the cell type-specific virus release defect in HeLa versus T cells, transient heterokaryons were produced between HeLa cells and the Jurkat T-cell line. These heterokaryons display a T-cell-like phenotype with respect to the requirement for p6 in particle release. The results described here define the role of p6 in virus replication in a wide range of cell types and reveal a strong cell type-dependent requirement for p6 in virus particle budding.
Figures




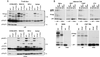



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

