Assembly of the RAG1/RAG2 synaptic complex
- PMID: 11739723
- PMCID: PMC134220
- DOI: 10.1128/MCB.22.1.69-77.2002
Assembly of the RAG1/RAG2 synaptic complex
Abstract
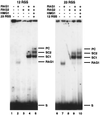
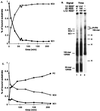
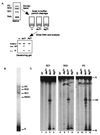
Assembly of antigen receptor genes by V(D)J recombination requires the site-specific recognition of two distinct DNA elements differing in the length of the spacer DNA that separates two conserved recognition motifs. Under appropriate conditions, V(D)J cleavage by the purified RAG1/RAG2 recombinase is similarly restricted. Double-strand breakage occurs only when these proteins are bound to a pair of complementary signals in a synaptic complex. We examine here the binding of the RAG proteins to signal sequences and find that the full complement of proteins required for synapsis of two signals and coupled cleavage can assemble on a single signal. This complex, composed of a dimer of RAG2 and at least a trimer of RAG1, remains inactive for double-strand break formation until a second complementary signal is provided. Thus, binding of the second signal activates the complex, possibly by inducing a conformational change. If synaptic complexes are formed similarly in vivo, one signal of a recombining pair may be the preferred site for RAG1/RAG2 assembly.
Figures



References
-
- Aldaz, H., E. Schuster, and T. A. Baker. 1996. The interwoven architecture of the Mu transposase couples DNA synapsis to catalysis. Cell 85:257–269. - PubMed
-
- Asante-Appiah, E., and A. M. Skalka. 1997. A metal-induced conformational change and activation of HIV-1 integrase. J. Biol. Chem. 272:16196–16205. - PubMed
-
- Baker, T. A., and K. Mizuuchi. 1992. DNA-promoted assembly of the active tetramer of the Mu transposase. Genes Dev. 6:2221–2232 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
