Gab3, a new DOS/Gab family member, facilitates macrophage differentiation
- PMID: 11739737
- PMCID: PMC134230
- DOI: 10.1128/MCB.22.1.231-244.2002
Gab3, a new DOS/Gab family member, facilitates macrophage differentiation
Abstract
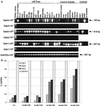
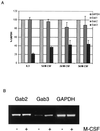
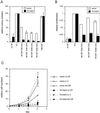
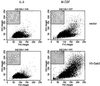
Using the FDC-P1 cell line expressing the exogenous macrophage colony-stimulating factor (M-CSF) receptor, Fms, we have analyzed the role of a new mammalian DOS/Gab-related signaling protein, called Gab3, in macrophage cell development of the mouse. Gab3 contains an amino-terminal pleckstrin homology domain, multiple potential sites for tyrosine phosphorylation and SH2 domain binding, and two major polyproline motifs potentially interacting with SH3 domains. Among the growing family of Gab proteins, Gab3 exhibits a unique and overlapping pattern of expression in tissues of the mouse compared with Gab1 and Gab2. Gab3 is more restricted to the hematopoietic tissues such as spleen and thymus but is detectable at progressively lower levels within heart, kidney, uterus, and brain. Like Gab2, Gab3 is tyrosine phosphorylated after M-CSF receptor stimulation and associates transiently with the SH2 domain-containing proteins p85 and SHP2. Overexpression of exogenous Gab3 in FD-Fms cells dramatically accelerates macrophage differentiation upon M-CSF stimulation. Unlike Gab2, which shows a constant mRNA expression level after M-CSF stimulation, Gab3 expression is initially absent or low in abundance in FD cells expressing the wild-type Fms, but Gab3 mRNA levels are increased upon M-CSF stimulation. Moreover, M-CSF stimulation of FD-FmsY807F cells (which grow but do not differentiate) fails to increase Gab3 expression. These results suggest that Gab3 is important for macrophage differentiation and that differentiation requires the early phosphorylation of Gab2 followed by induction and subsequent phosphorylation of Gab3.
Figures





References
-
- Allard, J. D., H. C. Chang, R. Herbst, H. McNeill, and M. A. Simon. 1996. The SH2-containing tyrosine phosphatase corkscrew is required during signaling by sevenless, Ras1 and Raf. Development 122:1137–1146. - PubMed
-
- Bardelli, A., P. Longati, D. Gramaglia, M. C. Stella, and P. M. Comoglio. 1997. Gab1 coupling to the HGF/Met receptor multifunctional docking site requires binding of Grb2 and correlates with the transforming potential. Oncogene 15:3103–3111. - PubMed
-
- Bausenwein, B. S., M. Schmidt, B. Mielke, and T. Raabe. 2000. In vivo functional analysis of the daughter of sevenless protein in receptor tyrosine kinase signaling. Mech. Dev. 90:205–215. - PubMed
-
- Bourette, R. P., G. M. Myles, K. Carlberg, and L. R. Rohrschneider. 1995. Uncoupling of the proliferation and differentiation signals mediated by the murine macrophage colony-stimulating factor receptor expressed in myeloid FDC-P1 cells. Cell Growth Differ. 6:631–645. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
