Yeast hnRNP K-like genes are involved in regulation of the telomeric position effect and telomere length
- PMID: 11739741
- PMCID: PMC134203
- DOI: 10.1128/MCB.22.1.286-297.2002
Yeast hnRNP K-like genes are involved in regulation of the telomeric position effect and telomere length
Abstract
Mammalian heterogeneous nuclear ribonucleoprotein K (hnRNP K) is an RNA- and DNA-binding protein implicated in the regulation of gene expression processes. To better understand its function, we studied two Saccharomyces cerevisiae homologues of the human hnRNP K, PBP2 and HEK2 (heterogeneous nuclear RNP K-like gene). pbp2Delta and hek2Delta mutations inhibited expression of a marker gene that was inserted near telomere but not at internal chromosomal locations. The telomere proximal to the ectopic marker gene became longer, while most of the other telomeres were not altered in the double mutant cells. We provide evidence that telomere elongation might be the primary event that causes enhanced silencing of an adjacent reporter gene. The telomere lengthening could, in part, be explained by the inhibitory effect of hek2Delta mutation on the telomeric rapid deletion pathway. Hek2p was detected in a complex with chromosome regions proximal to the affected telomere, suggesting a direct involvement of this protein in telomere maintenance. These results identify a role for hnRNP K-like genes in the structural and functional organization of telomeric chromatin in yeast.
Figures
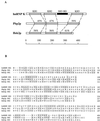






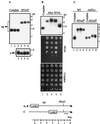

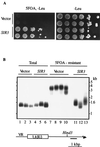
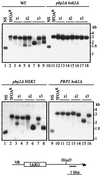

References
-
- Aparicio, O. M., B. L. Billington, and D. E. Gottschling. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66:1279–1287. - PubMed
-
- Baber, J. L., D. Libutti, D. Levens, and N. Tjandra. 1999. High precision solution structure of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K, a c-myc transcription factor. J. Mol. Biol. 289:949–962. - PubMed
-
- Bomsztyk, K., I. Van Seuningen, H. Suzuki, O. Denisenko, and J. Ostrowski. 1997. Diverse molecular interactions of the hnRNP K protein. FEBS Lett. 403:113–115. - PubMed
-
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132. - PubMed
-
- Burd, C. G., and G. Dreyfuss. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265:615–621. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
