Multiple roles of the tau131 subunit of yeast transcription factor IIIC (TFIIIC) in TFIIIB assembly
- PMID: 11739742
- PMCID: PMC134217
- DOI: 10.1128/MCB.22.1.298-308.2002
Multiple roles of the tau131 subunit of yeast transcription factor IIIC (TFIIIC) in TFIIIB assembly
Abstract
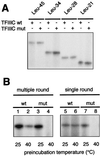
Yeast transcription factor IIIC (TFIIIC) plays a key role in assembling the transcription initiation factor TFIIIB on class III genes after TFIIIC-DNA binding. The second largest subunit of TFIIIC, tau131, is thought to initiate TFIIIB assembly by interacting with Brf1/TFIIIB70. In this work, we have analyzed a TFIIIC mutant (tau131-DeltaTPR2) harboring a deletion in tau131 removing the second of its 11 tetratricopeptide repeats. Remarkably, this thermosensitive mutation was selectively suppressed in vivo by overexpression of B"/TFIIIB90, but not Brf1 or TATA-binding protein. In vitro, the mutant factor preincubated at restrictive temperature bound DNA efficiently but lost transcription factor activity. The in vitro transcription defect was abolished at high concentrations of B" but not Brf1. Copurification experiments of baculovirus-expressed proteins confirmed a direct physical interaction between tau131 and B". tau131, therefore, appears to be involved in the recruitment of both Brf1 and B".
Figures







References
-
- Acker, J., M. de Graaff, I. Cheynel, V. Khazak, C. Kedinger, and M. Vigneron. 1997. Interactions between the human RNA polymerase II subunits. J. Biol. Chem. 272:16815–16821. - PubMed
-
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne, P. G. Amanatides, S. E. Scherer, P. W. Li, R. A. Hoskins, R. F. Galle, R. A. George, S. E. Lewis, S. Richards, M. Ashburner, S. N. Henderson, G. G. Sutton, J. R. Wortman, M. D. Yandell, Q. Zhang, L. X. Chen, R. C. Brandon, Y. H. Rogers, R. G. Blazej, M. Champe, B. D. Pfeiffer, K. H. Wan, C. Doyle, E. G. Baxter, G. Helt, C. R. Nelson, G. L. Gabor, J. F. Abril, A. Agbayani, H. J. An, C. Andrews-Pfannkoch, D. Baldwin, R. M. Ballew, A. Basu, J. Baxendale, L. Bayraktaroglu, E. M. Beasley, K. Y. Beeson, P. V. Benos, B. P. Berman, D. Bhandari, S. Bolshakov, D. Borkova, M. R. Botchan, J. Bouck, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185–2195. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
