Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell
- PMID: 11739776
- PMCID: PMC60751
- DOI: 10.1091/mbc.12.12.3733
Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell
Erratum in
-
Correction.Mol Biol Cell. 2021 Oct 1;32(20):cor1. doi: 10.1091/mbc.12.12.3733-corr. Mol Biol Cell. 2021. PMID: 34570654 Free PMC article. No abstract available.
Abstract
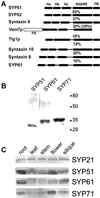
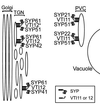
The syntaxin family of soluble N-ethyl maleimide sensitive factor adaptor protein receptors (SNAREs) is known to play an important role in the fusion of transport vesicles with specific organelles. Twenty-four syntaxins are encoded in the genome of the model plant Arabidopsis thaliana. These 24 genes are found in 10 gene families and have been reclassified as syntaxins of plants (SYPs). Some of these gene families have been previously characterized, with the SYP2-type syntaxins being found in the prevacuolar compartment (PVC) and the SYP4-type syntaxins on the trans-Golgi network (TGN). Here we report on two previously uncharacterized syntaxin groups. The SYP5 group is encoded by a two-member gene family, whereas SYP61 is a single gene. Both types of syntaxins are localized to multiple compartments of the endomembrane system, including the TGN and the PVC. These two groups of syntaxins form SNARE complexes with each other, and with other Arabidopsis SNAREs. On the TGN, SYP61 forms complexes with the SNARE VTI12 and either SYP41 or SYP42. SYP51 and SYP61 interact with each other and with VTI12, most likely also on the TGN. On the PVC, a SYP5-type syntaxin interacts specifically with a SYP2-type syntaxin, as well as the SNARE VTI11, forming a SNARE complex likely involved in TGN-to-PVC trafficking.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

