HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins
- PMID: 11739805
- PMCID: PMC60780
- DOI: 10.1091/mbc.12.12.4114
HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins
Abstract
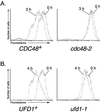
We isolated a temperature-sensitive mutant, hrd4-1, deficient in ER-associated degradation (ERAD). The HRD4 gene was identical to NPL4, a gene previously implicated in nuclear transport. Using a diverse set of substrates and direct ubiquitination assays, our analysis revealed that HRD4/NPL4 is required for a poorly characterized step in ERAD after ubiquitination of target proteins but before their recognition by the 26S proteasome. Our data indicate that this lack of proteasomal processing of ubiquitinated proteins constitutes the primary defect in hrd4/npl4 mutant cells and explains the diverse set of hrd4/npl4 phenotypes. We also found that each member of the Cdc48p-Ufd1p-Npl4p complex is individually required for ERAD.
Figures
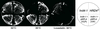





References
-
- Arias IM, Doyle D, Schimke RT. Studies on the synthesis and degradation of proteins of the endoplasmic reticulum of rat liver. J Biol Chem. 1969;244:3303–3315. - PubMed
-
- Bays NW, Gardner RG, Seelig LP, Joaezerio C, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. - PubMed
-
- Brodsky JL, McCracken AA. ER protein quality control and proteasome-mediated protein degradation. Semin Cell Dev Biol. 1999;10:507–513. - PubMed
-
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple Ubiquitin-Conjugating Enzymes Participate in the In Vivo Degradation of the Yeast Mat-α-2 Repressor. Cell. 1993;74:357–369. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

