AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching
- PMID: 11740565
- PMCID: PMC4729367
- DOI: 10.1038/414660a
AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching
Abstract
Class switch recombination (CSR) is a region-specific DNA recombination reaction that replaces one immunoglobulin heavy-chain constant region (Ch) gene with another. This enables a single variable (V) region gene to be used in conjunction with different downstream Ch genes, each having a unique biological activity. The molecular mechanisms that mediate CSR have not been defined, but activation-induced cytidine deaminase (AID), a putative RNA-editing enzyme, is required for this reaction. Here we report that the Nijmegen breakage syndrome protein (Nbs1) and phosphorylated H2A histone family member X (gamma-H2AX, also known as gamma-H2afx), which facilitate DNA double-strand break (DSB) repair, form nuclear foci at the Ch region in the G1 phase of the cell cycle in cells undergoing CSR, and that switching is impaired in H2AX-/- mice. Localization of Nbs1 and gamma-H2AX to the Igh locus during CSR is dependent on AID. In addition, AID is required for induction of switch region (S mu)-specific DNA lesions that precede CSR. These results place AID function upstream of the DNA modifications that initiate CSR.
Figures
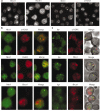


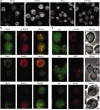
References
-
- Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. - PubMed
-
- Carney JP, et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. - PubMed
-
- Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. - PubMed
-
- Revy P, et al. Activation-induced cytidine deaminase (AID) de®ciency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 2000;102:565–575. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

