ATP-binding cassette transport system involved in regulation of morphological differentiation in response to glucose in Streptomyces griseus
- PMID: 11741848
- PMCID: PMC134767
- DOI: 10.1128/JB.184.1.91-103.2002
ATP-binding cassette transport system involved in regulation of morphological differentiation in response to glucose in Streptomyces griseus
Abstract
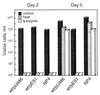
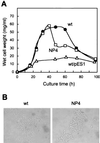
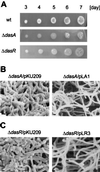
Streptomyces griseus NP4, which was derived by UV mutagenesis from strain IFO13350, showed a bald and wrinkled colony morphology in response to glucose. Mutant NP4 formed ectopic septa at intervals along substrate hyphae, and each of the compartments developed into a spore which was indistinguishable from an aerial spore in size, shape, and thickness of the spore wall and in susceptibility to lysozyme and heat. The ectopic spores of NP4 formed in liquid medium differed from "submerged spores" in lysozyme sensitivity. Shotgun cloning experiments with a library of the chromosomal DNA of the parental strain and mutant NP4 as the host gave rise to DNA fragments giving two different phenotypes; one complementing the bald phenotype of the host, and the other causing much severe wrinkled morphology in the host. Subcloning identified a gene (dasR) encoding a transcriptional repressor belonging to the GntR family that was responsible for the reversal of the bald phenotype and a gene (dasA) encoding a lipoprotein probably serving as a substrate-binding protein in an ATP-binding cassette (ABC) transport system that was responsible for the severe wrinkled morphology. These genes were adjacent but divergently encoded. Two genes, named dasB and dasC, encoding a membrane-spanning protein were present downstream of dasA, which suggested that dasRABC comprises a gene cluster for an ABC transporter, probably for sugar import. dasR was transcribed actively during vegetative growth, and dasA was transcribed just after commencement of aerial hypha formation and during sporulation, indicating that both were developmentally regulated. Transcriptional analysis and direct sequencing of dasRA in mutant NP4 suggested a defect of this mutant in the regulatory system to control the expression of these genes. Introduction of multicopies of dasA into the wild-type strain caused ectopic septation in very young substrate hyphae after only 1 day of growth and subsequent sporulation in response to glucose. The ectopic spores of the wild type had a thinner wall than those of mutant NP4, in agreement with the observation that the former was sensitive to lysozyme and heat. Disruption of the chromosomal dasA or dasR in the wild-type strain resulted in growth as substrate mycelium, suggesting an additional role of these genes in aerial mycelium formation. The ectopic septation and sporulation in mutant NP4 and the wild-type strain carrying multicopies of dasA were independent of a microbial hormone, A-factor (2-isocapryloyl-3R-hydroxymethyl-gamma-butyrolactone), that acts as a master switch of aerial mycelium formation and secondary metabolism.
Figures






References
-
- Amado, M., R. Almeida, T. Schwientek, and H. Clausen. 1999. Identification and characterization of large galactosyltransferase gene families: galactosyltransferases for all functions. Biochim. Biophys. Acta 1473:35–53. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingstone, D. O. Moore, J. S. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
-
- Beck, E., G. Ludwig, A. Auerswald, B. Reiss, and H. Schaller. 1982. Nucleotide sequence and exact localisation of the neomycin phosphotransferase gene from transposon Tn5. Gene 19:327–336. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

