IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins
- PMID: 11742080
- PMCID: PMC64955
- DOI: 10.1073/pnas.251550898
IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins
Abstract
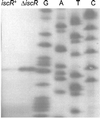
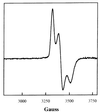
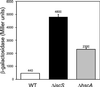
IscR (iron-sulfur cluster regulator) is encoded by an ORF located immediately upstream of genes coding for the Escherichia coli Fe-S cluster assembly proteins, IscS, IscU, and IscA. IscR shares amino acid similarity with MarA, a member of the MarA/SoxS/Rob family of transcription factors. In this study, we found that IscR functions as a repressor of the iscRSUA operon, because strains deleted for iscR have increased expression of this operon. In addition, in vitro transcription reactions established a direct role for IscR in repression of the iscR promoter. Analysis of IscR by electron paramagnetic resonance showed that the anaerobically isolated protein contains a [2Fe-2S](1+) cluster. The Fe-S cluster appears to be important for IscR function, because repression of iscR expression is significantly reduced in strains containing null mutations of the Fe-S cluster assembly genes iscS or hscA. The finding that IscR activity is decreased in strain backgrounds in which Fe-S cluster assembly is impaired suggests that this protein may be part of a novel autoregulatory mechanism that senses the Fe-S cluster assembly status of cells.
Figures




Comment in
-
Feedback regulation of iron-sulfur cluster biosynthesis.Proc Natl Acad Sci U S A. 2001 Dec 18;98(26):14751-3. doi: 10.1073/pnas.011579098. Proc Natl Acad Sci U S A. 2001. PMID: 11752417 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

