The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4
- PMID: 11742990
- PMCID: PMC125774
- DOI: 10.1093/emboj/20.24.7137
The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4
Abstract
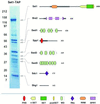
The SET domain proteins, SUV39 and G9a have recently been shown to be histone methyltransferases specific for lysines 9 and 27 (G9a only) of histone 3 (H3). The SET domains of the Saccharomyces cerevisiae Set1 and Drosophila trithorax proteins are closely related to each other but distinct from SUV39 and G9a. We characterized the complex associated with Set1 and Set1C and found that it is comprised of eight members, one of which, Bre2, is homologous to the trithorax-group (trxG) protein, Ash2. Set1C requires Set1 for complex integrity and mutation of Set1 and Set1C components shortens telomeres. One Set1C member, Swd2/Cpf10 is also present in cleavage polyadenylation factor (CPF). Set1C methylates lysine 4 of H3, thus adding a new specificity and a new subclass of SET domain proteins known to methyltransferases. Since methylation of H3 lysine 4 is widespread in eukaryotes, we screened the databases and found other Set1 homologues. We propose that eukaryotic Set1Cs are H3 lysine 4 methyltransferases and are related to trxG action through association with Ash2 homologues.
Figures









References
-
- Aasland R., Gibson,T.J. and Stewart,A.F. (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. - PubMed
-
- Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

