Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis
- PMID: 11743105
- PMCID: PMC133565
- DOI: 10.1104/pp.010467
Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis
Abstract
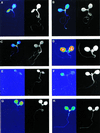
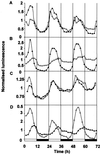
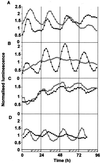
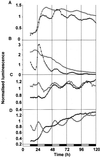
Many physiological and biochemical processes in plants exhibit endogenous rhythms with a period of about 24 h. Endogenous oscillators called circadian clocks regulate these rhythms. The circadian clocks are synchronized to the periodic environmental changes (e.g. day/night cycles) by specific stimuli; among these, the most important is the light. Photoreceptors, phytochromes, and cryptochromes are involved in setting the clock by transducing the light signal to the central oscillator. In this work, we analyzed the spatial, temporal, and long-term light-regulated expression patterns of the Arabidopsis phytochrome (PHYA to PHYE) and cryptochrome (CRY1 and CRY2) promoters fused to the luciferase (LUC(+)) reporter gene. The results revealed new details of the tissue-specific expression and light regulation of the PHYC and CRY1 and 2 promoters. More importantly, the data obtained demonstrate that the activities of the promoter::LUC(+) constructs, with the exception of PHYC::LUC(+), display circadian oscillations under constant conditions. In addition, it is shown by measuring the mRNA abundance of PHY and CRY genes under constant light conditions that the circadian control is also maintained at the level of mRNA accumulation. These observations indicate that the plant circadian clock controls the expression of these photoreceptors, revealing the formation of a new regulatory loop that could modulate gating and resetting of the circadian clock.
Figures

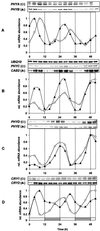


References
-
- Adam E, Szell M, Szekeres M, Schäfer E, Nagy F. The developmental and tissue-specific expression of tobacco phytochrome A genes. Plant J. 1994;6:283–293.
-
- Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of blue-light photoreceptor. Nature. 1993;366:162–166. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

