Negative regulation of phosphate starvation-induced genes
- PMID: 11743129
- PMCID: PMC133589
Negative regulation of phosphate starvation-induced genes
Abstract
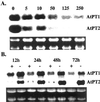
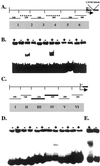
Phosphate (Pi) deficiency is a major nutritional problem faced by plants in many agro-ecosystems. This deficiency results in altered gene expression leading to physiological and morphological changes in plants. Altered gene expression is presumed to be due to interaction of regulatory sequences (cis-elements) present in the promoters with DNA binding factors (trans-factors). In this study, we analyzed the expression and DNA-protein interaction of promoter regions of Pi starvation-induced genes AtPT2 and TPSI1. AtPT2 encodes the high-affinity Pi transporter in Arabidopsis, whereas TPSI1 codes for a novel gene induced in the Pi-starved tomato (Lycopersicon esculentum). Expression of AtPT2 was induced rapidly under Pi deficiency and increased with decreasing concentrations of Pi. Abiotic stresses except Pi starvation had no affect on the expression of TPSI1. DNA mobility-shift assays indicated that specific sequences of AtPT2 and TPSI1 promoter interact with nuclear protein factors. Two regions of AtPT2 and TPSI1 promoters specifically bound nuclear protein factors from Pi-sufficient plants. Interestingly, the DNA binding activity disappeared during Pi starvation, leading to the hypothesis that Pi starvation-induced genes may be under negative regulation.
Figures




Similar articles
-
Differential expression of TPS11, a phosphate starvation-induced gene in tomato.Plant Mol Biol. 1997 Mar;33(5):867-74. doi: 10.1023/a:1005729309569. Plant Mol Biol. 1997. PMID: 9106510
-
Phosphite, an analog of phosphate, suppresses the coordinated expression of genes under phosphate starvation.Plant Physiol. 2002 Jul;129(3):1232-40. doi: 10.1104/pp.010835. Plant Physiol. 2002. PMID: 12114577 Free PMC article.
-
Brassica napus PHR1 gene encoding a MYB-like protein functions in response to phosphate starvation.PLoS One. 2012;7(8):e44005. doi: 10.1371/journal.pone.0044005. Epub 2012 Aug 29. PLoS One. 2012. PMID: 22952851 Free PMC article.
-
A Brassica napus PHT1 phosphate transporter, BnPht1;4, promotes phosphate uptake and affects roots architecture of transgenic Arabidopsis.Plant Mol Biol. 2014 Dec;86(6):595-607. doi: 10.1007/s11103-014-0249-y. Epub 2014 Sep 7. Plant Mol Biol. 2014. PMID: 25194430
-
Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation.Plant Physiol. 2014 Apr;164(4):2020-9. doi: 10.1104/pp.113.235077. Epub 2014 Feb 28. Plant Physiol. 2014. PMID: 24586044 Free PMC article.
Cited by
-
Promoter analysis of the barley Pht1;1 phosphate transporter gene identifies regions controlling root expression and responsiveness to phosphate deprivation.Plant Physiol. 2004 Dec;136(4):4205-14. doi: 10.1104/pp.104.045823. Epub 2004 Nov 12. Plant Physiol. 2004. PMID: 15542491 Free PMC article.
-
Beyond the proteome: non-coding regulatory RNAs.Genome Biol. 2002;3(5):reviews0005. doi: 10.1186/gb-2002-3-5-reviews0005. Epub 2002 Apr 15. Genome Biol. 2002. PMID: 12049667 Free PMC article. Review.
-
Purple acid phosphatases: roles in phosphate utilization and new emerging functions.Plant Cell Rep. 2022 Jan;41(1):33-51. doi: 10.1007/s00299-021-02773-7. Epub 2021 Aug 17. Plant Cell Rep. 2022. PMID: 34402946 Review.
-
Medicago truncatula PHO2 genes have distinct roles in phosphorus homeostasis and symbiotic nitrogen fixation.Front Plant Sci. 2023 Jun 13;14:1211107. doi: 10.3389/fpls.2023.1211107. eCollection 2023. Front Plant Sci. 2023. PMID: 37409286 Free PMC article.
-
Early Transcriptomic Response to Phosphate Deprivation in Soybean Leaves as Revealed by RNA-Sequencing.Int J Mol Sci. 2018 Jul 23;19(7):2145. doi: 10.3390/ijms19072145. Int J Mol Sci. 2018. PMID: 30041471 Free PMC article.
References
-
- Barber SA. Soil-plant interactions in the phosphorus nutrition of plants. In: Khasawneh FE, Sample EC, Kamprath EJ, editors. Role of Phosphorus in Agriculture. Madison, WI: American Society of Agronomy; 1980. pp. 591–615.
-
- Biddinger EC, Liu C, Joly RJ, Raghothama KG. Physiological and molecular responses of aeroponically grown tomato plants to phosphorus deficiency. J Am Soc Hortic Sci. 1998;123:330–333.
-
- Blackwell TK, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. - PubMed
-
- Burleigh SH, Harrison MJ. A novel gene whose expression in Medicago truncatularoots is suppressed in response to colonization by vesicular-arbuscular mycorrhizal (VAM) fungi and to phosphate nutrition. Plant Mol Biol. 1997;34:199–208. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
