Genomic approach for analysis of surface proteins in Chlamydia pneumoniae
- PMID: 11748203
- PMCID: PMC127649
- DOI: 10.1128/IAI.70.1.368-379.2002
Genomic approach for analysis of surface proteins in Chlamydia pneumoniae
Abstract
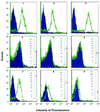
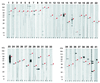
Chlamydia pneumoniae, a human pathogen causing respiratory infections and probably contributing to the development of atherosclerosis and heart disease, is an obligate intracellular parasite which for replication needs to productively interact with and enter human cells. Because of the intrinsic difficulty in working with C. pneumoniae and in the absence of reliable tools for its genetic manipulation, the molecular definition of the chlamydial cell surface is still limited, thus leaving the mechanisms of chlamydial entry largely unknown. In an effort to define the surface protein organization of C. pneumoniae, we have adopted a combined genomic-proteomic approach based on (i) in silico prediction from the available genome sequences of peripherally located proteins, (ii) heterologous expression and purification of selected proteins, (iii) production of mouse immune sera against the recombinant proteins to be used in Western blotting and fluorescence-activated cell sorter (FACS) analyses for the identification of surface antigens, and (iv) mass spectrometry analysis of two-dimensional electrophoresis (2DE) maps of chlamydial protein extracts to confirm the presence of the FACS-positive antigens in the chlamydial cell. Of the 53 FACS-positive sera, 41 recognized a protein species with the expected size on Western blots, and 28 of the 53 antigens shown to be surface-exposed by FACS were identified on 2DE maps of elementary-body extracts. This work represents the first systematic attempt to define surface protein organization in C. pneumoniae.
Figures

References
-
- Bavoil, P. M., R. C. Hsia, and D. Ojcius. 2000. Closing in on Chlamydia and its intracellular bag of tricks. Microbiology 146: 2723–2731. - PubMed
-
- Bavoil, P. M., and R. C Hsia. 1998. Type III secretion in Chlamydia: a case of deja vu? Mol. Microbiol. 28: 860–862. - PubMed
-
- Bjellqvist, B., G. J. Hughes, C. Pasquali, N. Paquet, F. Ravier, J. C. Sanchez, S. Frutiger, and D. Hochstrasser. 1993. The focusing positions of poly-peptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14: 1023–1031. - PubMed
-
- Chevallet, M., V. Santoni, A. Poinas, D. Rouquie, A. Fuchs, S. Kieffer, M. Rossignol, J. Lunardi, J. Garin, and T. Rabilloud. 1998. New zwitterionic detergents improve the analysis of membrane proteins by two-dimensional electrophoresis. Electrophoresis 19: 1901–1909. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

