Mycobacterium smegmatis D-Alanine Racemase Mutants Are Not Dependent on D-Alanine for Growth
- PMID: 11751110
- PMCID: PMC126997
- DOI: 10.1128/AAC.46.2.47-54.2002
Mycobacterium smegmatis D-Alanine Racemase Mutants Are Not Dependent on D-Alanine for Growth
Abstract
Mycobacterium smegmatis is a fast-growing nonpathogenic species particularly useful in studying basic cellular processes of relevance to pathogenic mycobacteria. This study focused on the D-alanine racemase gene (alrA), which is involved in the synthesis of D-alanine, a basic component of peptidoglycan that forms the backbone of the cell wall. M. smegmatis alrA null mutants were generated by homologous recombination using a kanamycin resistance marker for insertional inactivation. Mutants were selected on Middlebrook medium supplemented with 50 mM D-alanine and 20 microg of kanamycin per ml. These mutants were also able to grow in standard and minimal media without D-alanine, giving rise to colonies with a drier appearance and more-raised borders than the wild-type strain. The viability of the mutants and independence of D-alanine for growth indicate that inactivation of alrA does not impose an auxotrophic requirement for D-alanine, suggesting the existence of a new pathway of D-alanine biosynthesis in M. smegmatis. Biochemical analysis demonstrated the absence of any detectable D-alanine racemase activity in the mutant strains. In addition, the alrA mutants displayed hypersusceptibility to the antimycobacterial agent D-cycloserine. The MIC of D-cycloserine for the mutant strain was 2.56 microg/ml, 30-fold less than that for the wild-type strain. Furthermore, this hypersusceptibility was confirmed by the bactericidal action of D-cycloserine on broth cultures. The kinetic of killing for the mutant strain followed the same pattern as that for the wild-type strain, but at a 30-fold-lower drug concentration. This effect does not involve a change in the permeability of the cell wall by this drug and is consistent with the identification of D-alanine racemase as a target of D-cycloserine. This outcome is of importance for the design of novel antituberculosis drugs targeting peptidoglycan biosynthesis in mycobacteria.
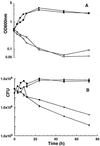
Figures


Similar articles
-
Overexpression of the D-alanine racemase gene confers resistance to D-cycloserine in Mycobacterium smegmatis.J Bacteriol. 1997 Aug;179(16):5046-55. doi: 10.1128/jb.179.16.5046-5055.1997. J Bacteriol. 1997. PMID: 9260945 Free PMC article.
-
Use of NMR metabolomics to analyze the targets of D-cycloserine in mycobacteria: role of D-alanine racemase.J Proteome Res. 2007 Dec;6(12):4608-14. doi: 10.1021/pr0704332. Epub 2007 Nov 3. J Proteome Res. 2007. PMID: 17979227 Free PMC article.
-
The alanine racemase of Mycobacterium smegmatis is essential for growth in the absence of D-alanine.J Bacteriol. 2007 Nov;189(22):8381-6. doi: 10.1128/JB.01201-07. Epub 2007 Sep 7. J Bacteriol. 2007. PMID: 17827284 Free PMC article.
-
Inhibitors of alanine racemase enzyme: a review.J Enzyme Inhib Med Chem. 2016 Aug;31(4):517-26. doi: 10.3109/14756366.2015.1050010. Epub 2015 May 29. J Enzyme Inhib Med Chem. 2016. PMID: 26024289 Review.
-
Molecular basis underlying Mycobacterium tuberculosis D-cycloserine resistance. Is there a role for ubiquinone and menaquinone metabolic pathways?Expert Opin Ther Targets. 2014 Jun;18(6):691-701. doi: 10.1517/14728222.2014.902937. Epub 2014 Apr 29. Expert Opin Ther Targets. 2014. PMID: 24773568 Review.
Cited by
-
Generation and screening of a comprehensive Mycobacterium avium subsp. paratuberculosis transposon mutant bank.Front Cell Infect Microbiol. 2014 Oct 15;4:144. doi: 10.3389/fcimb.2014.00144. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25360421 Free PMC article.
-
Identifying feasible metabolic routes in Mycobacterium smegmatis and possible alterations under diverse nutrient conditions.BMC Microbiol. 2014 Nov 18;14:276. doi: 10.1186/s12866-014-0276-5. BMC Microbiol. 2014. PMID: 25403821 Free PMC article.
-
Purification, Characterization and Inhibition of Alanine Racemase from a Pathogenic Strain of Streptococcus iniae.Pol J Microbiol. 2019 Sep;68(3):331-341. doi: 10.33073/pjm-2019-036. Epub 2019 Sep 3. Pol J Microbiol. 2019. PMID: 31880879 Free PMC article.
-
Assessment of Metabolic Changes in Mycobacterium smegmatis Wild-Type and alr Mutant Strains: Evidence of a New Pathway of d-Alanine Biosynthesis.J Proteome Res. 2017 Mar 3;16(3):1270-1279. doi: 10.1021/acs.jproteome.6b00871. Epub 2017 Feb 14. J Proteome Res. 2017. PMID: 28121156 Free PMC article.
-
A Kinetic Study of In Vitro Lysis of Mycobacterium smegmatis.Chem Eng Sci. 2009 May 1;64(9):1944-1952. doi: 10.1016/j.ces.2008.12.015. Chem Eng Sci. 2009. PMID: 21451732 Free PMC article.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1990. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
-
- Belanger, A. E., and J. M. Inamine. 2000. Genetics of cell wall biosynthesis, p.191–202. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

