Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae
- PMID: 11751634
- PMCID: PMC312847
- DOI: 10.1101/gad.940201
Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae
Abstract
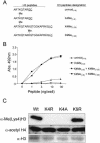
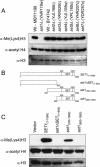
Histone methylation is known to be associated with both transcriptionally active and repressive chromatin states. Recent studies have identified SET domain-containing proteins such as SUV39H1 and Clr4 as mediators of H3 lysine 9 (Lys9) methylation and heterochromatin formation. Interestingly, H3 Lys9 methylation is not observed from bulk histones isolated from asynchronous populations of Saccharomyces cerevisiae or Tetrahymena thermophila. In contrast, H3 lysine 4 (Lys4) methylation is a predominant modification in these smaller eukaryotes. To identify the responsible methyltransferase(s) and to gain insight into the function of H3 Lys4 methylation, we have developed a histone H3 Lys4 methyl-specific antiserum. With this antiserum, we show that deletion of SET1, but not of other putative SET domain-containing genes, in S. cerevisiae, results in the complete abolishment of H3 Lys4 methylation in vivo. Furthermore, loss of H3 Lys4 methylation in a set1 Delta strain can be rescued by SET1. Analysis of histone H3 mutations at Lys4 revealed a slow-growth defect similar to a set1 Delta strain. Chromatin immunoprecipitation assays show that H3 Lys4 methylation is present at the rDNA locus and that Set1-mediated H3 Lys4 methylation is required for repression of RNA polymerase II transcription within rDNA. Taken together, these data suggest that Set1-mediated H3 Lys4 methylation is required for normal cell growth and transcriptional silencing.
Figures



References
-
- Aagaard L, Schmid M, Warburton P, Jenuwein T. Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J Cell Sci. 2000;113:817–829. - PubMed
-
- Annunziato AT, Eason MB, Perry CA. Relationship between methylation and acetylation of arginine-rich histones in cycling and arrested HeLa cells. Biochemistry. 1995;34:2916–2924. - PubMed
-
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
-
- Boggs, B.A., Cheung, P., Heard, E., Spector, D.L., Chinault, C., and Allis, C.D. 2001. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. (in press). - PubMed
-
- Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes & Dev. 1997;11:255–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
