A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers
- PMID: 11751640
- PMCID: PMC312850
- DOI: 10.1101/gad.931001
A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers
Erratum in
- Genes Dev 2002 Mar 15;16(6):764
Abstract
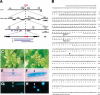
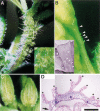
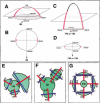
It is postulated that the symmetric organization of plant lateral organs is based on two crossed axes, the abaxial-adaxial and the lateral axes. The PRESSED FLOWER (PRS) gene, the expression and function of which are dependent on the lateral axis, is reported in this study. In the prs mutant, growth of the lateral sepals is repressed, and although the size and shape of the abaxial and adaxial sepals are normal, the cell files at the lateral margins are missing. Double-mutant analyses showed that the PRS gene functions independently of the determinations of both floral organ identity and floral meristem size. The PRS gene, encoding a putative transcriptional factor with a homeodomain, was shown to be required for cell proliferation. PRS gene expression is spatially and temporally unique and is expressed in a restricted number of L1 cells at the lateral regions of flower primordia, floral organ primordia, and young leaf primordia. Our study strongly suggests that the PRS gene is involved in the molecular mechanism of lateral axis-dependent development of lateral organs in Arabidopsis.
Figures


References
-
- Abe M, Takahashi T, Komeda Y. Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 2001;26:487–494. - PubMed
-
- Bossinger G, Smyth DR. Initiation patterns of flower and floral organ development in Arabidopsis thaliana. Development. 1996;122:1093–1102. - PubMed
-
- Bowman JL. Morphology of the expanded first leaves. In: Bowman JL, editor. Arabidopsis: An atlas of morphology and development. New York, NY: Springer-Verlag; 1994. pp. 38–39.
-
- ————— Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
