Characterization of comQ and comX, two genes required for production of ComX pheromone in Bacillus subtilis
- PMID: 11751817
- PMCID: PMC139578
- DOI: 10.1128/JB.184.2.410-419.2002
Characterization of comQ and comX, two genes required for production of ComX pheromone in Bacillus subtilis
Abstract
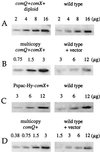
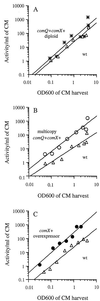
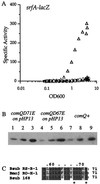
Many microbes use secreted peptide-signaling molecules to stimulate changes in gene expression in response to high population density, a process called quorum sensing. ComX pheromone is a modified 10-amino-acid peptide used by Bacillus subtilis to modulate changes in gene expression in response to crowding. comQ and comX are required for production of ComX pheromone. We found that accumulation of ComX pheromone in culture supernatant paralleled cell growth, indicating that there was no autoinduction of production of ComX pheromone. We overexpressed comQ and comX separately and together and found that overexpression of comX alone was sufficient to cause an increase in production of ComX pheromone and early induction of a quorum-responsive promoter. These results indicate that the extracellular concentration of ComX pheromone plays a major role in determining the timing of the quorum response and that expression of comX is limiting for production of ComX pheromone. We made alanine substitutions in the residues that comprise the peptide backbone of ComX pheromone. Analysis of these mutants highlighted the importance of the modification for ComX pheromone function and identified three residues (T50, G54, and D55) that are unlikely to interact with proteins involved in production of or response to ComX pheromone. We have also identified and mutated a putative isoprenoid binding domain of ComQ. Mutations in this domain eliminated production of ComX pheromone, consistent with the hypothesis that ComQ is involved in modifying ComX pheromone and that the modification is likely to be an isoprenoid.
Figures




References
-
- Ashby, M. N., D. H. Spear, and P. A. Edwards. 1990. Prenyltransferases: from yeast to man, p27–34. In A. D. Attie (ed.), Molecular biology of atherosclerosis. Elsevier, Inc., New York, N.Y.
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1990. Current protocols in molecular biology. Greene Publishing Associates and Wiley Interscience, New York, N.Y.
-
- Caldwell, G. A., S. H. Wang, C. B. Xue, Y. Jiang, H. F. Lu, F. Naider, and J. M. Becker. 1994. Molecular determinants of bioactivity of the Saccharomyces cerevisiae lipopeptide mating pheromone. J. Biol. Chem. 269:19817–19825. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

