Bacillus subtilis tolerance of moderate concentrations of rifampin involves the sigma(B)-dependent general and multiple stress response
- PMID: 11751823
- PMCID: PMC139561
- DOI: 10.1128/JB.184.2.459-467.2002
Bacillus subtilis tolerance of moderate concentrations of rifampin involves the sigma(B)-dependent general and multiple stress response
Abstract
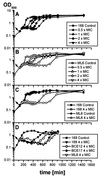
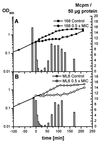
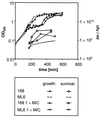
Low concentrations of the RNA polymerase inhibitor rifampin added to an exponentially growing culture of Bacillus subtilis led to an instant inhibition of growth. Survival experiments revealed that during the growth arrest the cells became tolerant to the antibiotic and the culture was able to resume growth some time after rifampin treatment. L-[(35)S]methionine pulse-labeled protein extracts were separated by two-dimensional polyacrylamide gel electrophoresis to investigate the change in the protein synthesis pattern in response to rifampin. The sigma(B)-dependent general stress proteins were found to be induced after treatment with the antibiotic. Part of the oxidative stress signature was induced as indicated by the catalase KatA and MrgA. The target protein of rifampin, the beta subunit (RpoB) of the DNA-dependent RNA polymerase, and the flagellin protein Hag belonging to the sigma(D) regulon were also induced. The rifampin-triggered growth arrest was extended in a sigB mutant in comparison to the wild-type strain, and the higher the concentration, the more pronounced this effect was. Activity of the RsbP energy-signaling phosphatase in the sigma(B) signal transduction network was also important for this protection against rifampin, but the RsbU environmental signaling phosphatase was not required. The sigB mutant strain was less capable of growing on rifampin-containing agar plates. When plated from a culture that had already reached stationary phase without previous exposure to the antibiotic during growth, the survival rate of the wild type exceeded that of the sigB mutant by a factor of 100. We conclude that the general stress response of B. subtilis is induced by rifampin depending on RsbP activity and that loss of SigB function causes increased sensitivity to the antibiotic.
Figures





References
-
- Bernhardt, J., K. Büttner, C. Scharf, and M. Hecker. 1999. Dual channel imaging of two-dimensional electropherograms in Bacillus subtilis. Electrophoresis 20:2225–2240. - PubMed
-
- Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901–912. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

