Sequential partially overlapping gene arrangement in the tricistronic S1 genome segments of avian reovirus and Nelson Bay reovirus: implications for translation initiation
- PMID: 11752152
- PMCID: PMC136829
- DOI: 10.1128/jvi.76.2.609-618.2002
Sequential partially overlapping gene arrangement in the tricistronic S1 genome segments of avian reovirus and Nelson Bay reovirus: implications for translation initiation
Abstract
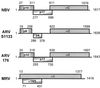
Previous studies of the avian reovirus strain S1133 (ARV-S1133) S1 genome segment revealed that the open reading frame (ORF) encoding the final sigmaC viral cell attachment protein initiates over 600 nucleotides distal from the 5' end of the S1 mRNA and is preceded by two predicted small nonoverlapping ORFs. To more clearly define the translational properties of this unusual polycistronic RNA, we pursued a comparative analysis of the S1 genome segment of the related Nelson Bay reovirus (NBV). Sequence analysis indicated that the 3'-proximal ORF present on the NBV S1 genome segment also encodes a final sigmaC homolog, as evidenced by the presence of an extended N-terminal heptad repeat characteristic of the coiled-coil region common to the cell attachment proteins of reoviruses. Most importantly, the NBV S1 genome segment contains two conserved ORFs upstream of the final sigmaC coding region that are extended relative to the predicted ORFs of ARV-S1133 and are arranged in a sequential, partially overlapping fashion. Sequence analysis of the S1 genome segments of two additional strains of ARV indicated a similar overlapping tricistronic gene arrangement as predicted for the NBV S1 genome segment. Expression analysis of the ARV S1 genome segment indicated that all three ORFs are functional in vitro and in virus-infected cells. In addition to the previously described p10 and final sigmaC gene products, the S1 genome segment encodes from the central ORF a 17-kDa basic protein (p17) of no known function. Optimizing the translation start site of the ARV p10 ORF lead to an approximately 15-fold increase in p10 expression with little or no effect on translation of the downstream final sigmaC ORF. These results suggest that translation initiation complexes can bypass over 600 nucleotides and two functional overlapping upstream ORFs in order to access the distal final sigmaC start site.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

