Evidence for human immunodeficiency virus type 1 replication in vivo in CD14(+) monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy
- PMID: 11752161
- PMCID: PMC136806
- DOI: 10.1128/jvi.76.2.707-716.2002
Evidence for human immunodeficiency virus type 1 replication in vivo in CD14(+) monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy
Abstract
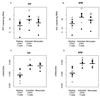
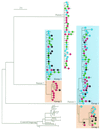
In vitro studies show that human immunodeficiency virus type 1 (HIV-1) does not replicate in freshly isolated monocytes unless monocytes differentiate to monocyte-derived macrophages. Similarly, HIV-1 may replicate in macrophages in vivo, whereas it is unclear whether blood monocytes are permissive to productive infection with HIV-1. We investigated HIV-1 replication in CD14(+) monocytes and resting and activated CD4(+) T cells by measuring the levels of cell-associated viral DNA and mRNA and the genetic evolution of HIV-1 in seven acutely infected patients whose plasma viremia had been <100 copies/ml for 803 to 1,544 days during highly active antiretroviral therapy (HAART). HIV-1 DNA was detected in CD14(+) monocytes as well as in activated and resting CD4(+) T cells throughout the course of study. While significant variation in the decay slopes of HIV-1 DNA was seen among individual patients, viral decay in CD14(+) monocytes was on average slower than that in activated and resting CD4(+) T cells. Measurements of HIV-1 sequence evolution and the concentrations of unspliced and multiply spliced mRNA provided evidence of ongoing HIV-1 replication, more pronounced in CD14(+) monocytes than in resting CD4(+) T cells. Phylogenetic analyses of HIV-1 sequences indicated that after prolonged HAART, viral populations related or identical to those found only in CD14(+) monocytes were seen in plasma from three of the seven patients. In the other four patients, HIV-1 sequences in plasma and the three cell populations were identical. CD14(+) monocytes appear to be one of the potential in vivo sources of HIV-1 in patients receiving HAART.
Figures



References
-
- Aquaro, S., C. F. Perno, E. Balestra, J. Balzarini, A. Cenci, A. Bertoli, M. Francesconi, S. Panti, F. Serra, N. Villani, and R. Calio. 1997. Inhibition of replication of HIV in primary monocyte/macrophages by different antiviral drugs and comparative efficacy in lymphocytes. J. Leukoc. Biol. 62:138–143. - PubMed
-
- Aquaro, S., R. Calio, E. Balestra, P. Bagnarelli, A. Cenci, A. Bertoli, B. Tavazzi, D. DiPierro, M. Francesconi, D. Abdelahad, and C. F. Perno. 1998. Clinical implications of HIV dynamics and drug resistance in macrophages. J. Biol. Regul. Homeost. Agents 12(Suppl. 1–2):23–27. - PubMed
-
- Berrey, M. M., T. Schacker, A. C. Collier, T. Shea, S. J. Brodie, D. Mayers, R. Coombs, J. Krieger, T. W. Chun, A. S. Fauci, S. G. Self, and L. Corey. 2001. Treatment of primary human immunodeficiency virus type 1 infection with potent antiretroviral therapy reduces frequency of rapid progression to AIDS. J. Infect. Dis. 183:1466–1475. - PubMed
-
- Blankson, J. N., D. Finzi, T. C. Pierson, B. P. Sabundayo, K. Chadwick, J. B. Margolick, T. C. Quinn, and R. F. Siliciano. 2000. Biphasic decay of latently infected CD4+ T cells in acute human immunodeficiency virus type 1 infection. J. Infect. Dis. 182:1636–1642. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

