K15 protein of Kaposi's sarcoma-associated herpesvirus is latently expressed and binds to HAX-1, a protein with antiapoptotic function
- PMID: 11752170
- PMCID: PMC136811
- DOI: 10.1128/jvi.76.2.802-816.2002
K15 protein of Kaposi's sarcoma-associated herpesvirus is latently expressed and binds to HAX-1, a protein with antiapoptotic function
Abstract
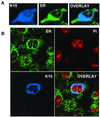
The Kaposi's sarcoma-associated herpesvirus (KSHV) (or human herpesvirus 8) open reading frame (ORF) K15 encodes a putative integral transmembrane protein in the same genomic location as latent membrane protein 2A of Epstein-Barr virus. Ectopic expression of K15 in cell lines revealed the presence of several different forms ranging in size from full length, approximately 50 kDa, to 17 kDa. Of these different species the 35- and 23-kDa forms were predominant. Mutational analysis of the initiator AUG indicated that translation initiation from this first AUG is required for K15 expression. Computational analysis indicates that the different forms detected may arise due to proteolytic cleavage at internal signal peptide sites. We show that K15 is latently expressed in KSHV-positive primary effusion lymphoma cell lines and in multicentric Castleman's disease. Using a yeast two-hybrid screen we identified HAX-1 (HS1 associated protein X-1) as a binding partner to the C terminus of K15 and show that K15 interacts with cellular HAX-1 in vitro and in vivo. Furthermore, HAX-1 colocalizes with K15 in the endoplasmic reticulum and mitochondria. The function of HAX-1 is unknown, although the similarity of its sequence to those of Nip3 and Bcl-2 infers a role in the regulation of apoptosis. We show here that HAX-1 can form homodimers in vivo and is a potent inhibitor of apoptosis and therefore represents a new apoptosis regulatory protein. The putative functions of K15 with respect to its interaction with HAX-1 are discussed.
Figures













References
-
- Boise, L. H., M. Gonzalez-Garcia, C. E. Postema, L. Ding, T. Lindsten, L. A. Turka, X. Mao, G. Nunez, and C. B. Thompson. 1993. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74:597–608. - PubMed
-
- Boshoff, C., and Y. Chang. 2001. Kaposi’s sarcoma-associated herpesvirus: a new DNA tumor virus. Annu. Rev. Med. 52:453–470. - PubMed
-
- Choi, J., R. E. Means, B. Damania, and J. U. Jung. 2001. Molecular piracy of Kaposi’s sarcoma associated herpesvirus. Cytokine Growth Factor Rev. 12:245–257. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

