PlasmoDB: the Plasmodium genome resource. An integrated database providing tools for accessing, analyzing and mapping expression and sequence data (both finished and unfinished)
- PMID: 11752262
- PMCID: PMC99106
- DOI: 10.1093/nar/30.1.87
PlasmoDB: the Plasmodium genome resource. An integrated database providing tools for accessing, analyzing and mapping expression and sequence data (both finished and unfinished)
Abstract
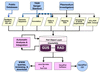
PlasmoDB (http://PlasmoDB.org) is the official database of the Plasmodium falciparum genome sequencing consortium. This resource incorporates finished and draft genome sequence data and annotation emerging from Plasmodium sequencing projects. PlasmoDB currently houses information from five parasite species and provides tools for cross-species comparisons. Sequence information is also integrated with other genomic-scale data emerging from the Plasmodium research community, including gene expression analysis from EST, SAGE and microarray projects. The relational schemas used to build PlasmoDB [Genomics Unified Schema (GUS) and RNA Abundance Database (RAD)] employ a highly structured format to accommodate the diverse data types generated by sequence and expression projects. A variety of tools allow researchers to formulate complex, biologically based queries of the database. A version of the database is also available on CD-ROM (Plasmodium GenePlot), facilitating access to the data in situations where Internet access is difficult (e.g. by malaria researchers working in the field). The goal of PlasmoDB is to enhance utilization of the vast quantities of data emerging from genome-scale projects by the global malaria research community.
Figures

References
-
- Fletcher C. (1998) The Plasmodium falciparum Genome Project. Parasitol. Today, 14, 342–344. - PubMed
-
- Davidson S., Crabtree,J., Brunk,B.P., Schug,J., Tannen,V., Overton,G.C. and Stoeckert,C.J.,Jr (2001) K2/Klesli and GUS: experiments in integrated access to genomic data sources. IBM Systems J., 40, 512–531.
-
- Stoeckert C., Pizarro,A., Manduchi,E., Gibson,M., Brunk,B., Crabtree,J., Schug,J., Shen-Orr,S. and Overton,C.G. (2001) A relational schema for both array-based and SAGE gene expression experiments. Bioinformatics, 17, 300–308. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

