The Celera Discovery System
- PMID: 11752274
- PMCID: PMC99167
- DOI: 10.1093/nar/30.1.129
The Celera Discovery System
Abstract
The Celera Discovery System (CDS) is a web-accessible research workbench for mining genomic and related biological information. Users have access to the human and mouse genome sequences with annotation presented in summary form in BioMolecule Reports for genes, transcripts and proteins. Over 40 additional databases are available, including sequence, mapping, mutation, genetic variation, mRNA expression, protein structure, motif and classification data. Data are accessible by browsing reports, through a variety of interactive graphical viewers, and by advanced query capability provided by the LION SRS search engine. A growing number of sequence analysis tools are available, including sequence similarity, pattern searching, multiple sequence alignment and Hidden Markov Model search. A user workspace keeps track of queries and analyses. CDS is widely used by the academic research community and requires a subscription for access. The system and academic pricing information are available at http://cds.celera.com.
Figures


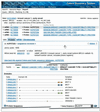
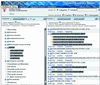

References
-
- Velculescu V.E., Zhang,L., Vogelstein,B. and Kinzler,K.W. (1995), Serial analysis of gene expression. Science, 270, 484–487. - PubMed
-
- Brenner S., Johnson,M., Bridgham,J., Golda,G., Lloyd,D.H., Johnson,D., Luo,S., McCurdy,S., Foy,M., Ewan,M. et. al. (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol., 18, 630–634. - PubMed
-
- Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et. al. (2001) The sequence of the human genome. Science, 291, 1304–1351. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

