HugeIndex: a database with visualization tools for high-density oligonucleotide array data from normal human tissues
- PMID: 11752297
- PMCID: PMC99144
- DOI: 10.1093/nar/30.1.214
HugeIndex: a database with visualization tools for high-density oligonucleotide array data from normal human tissues
Abstract
High-density oligonucleotide arrays are a powerful tool for uncovering changes in global gene expression in various disease states. To this end, it is essential to first characterize the variations of gene expression in normal physiological processes. We established the Human Gene Expression (HuGE) Index database (www.HugeIndex.org) to serve as a public repository for gene expression data on normal human tissues using high-density oligonucleotide arrays. This resource currently contains the results of 59 gene expression experiments on 19 human tissues. We provide interactive tools for researchers to query and visualize our data over the Internet. To facilitate data analysis, we cross-reference each gene on the array with its annotation in the LocusLink database at NCBI.
Figures

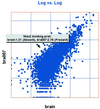

References
-
- Lockhart D.J. and Winzeler,E.A. (2000) Genomics, gene expression and DNA arrays. Nature, 405, 827–836. - PubMed
-
- DeRisi J., Penland,L., Brown,P.O., Bittner,M.L., Meltzer,P.S., Ray,M., Chen,Y., Su,Y.A. and Trent,J.M. (1996) Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nature Genet., 14, 457–460. - PubMed
-
- Golub T.R., Slonim,D.K., Tamayo,P., Huard,C., Gaasenbeek,M., Mesirov,J.P., Coller,H., Loh,M.L., Downing,J.R., Caligiuri,M.A. et al. (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science, 286, 531–537. - PubMed
-
- Alizadeh A.A., Eisen,M.B., Davis,R.E., Ma,C., Lossos,I.S., Rosenwald,A., Boldrick,J.C., Sabet,H., Tran,T., Yu,X. et al. (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature, 403, 503–511. - PubMed

