The Pfam protein families database
- PMID: 11752314
- PMCID: PMC99071
- DOI: 10.1093/nar/30.1.276
The Pfam protein families database
Abstract
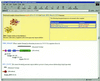
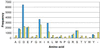
Pfam is a large collection of protein multiple sequence alignments and profile hidden Markov models. Pfam is available on the World Wide Web in the UK at http://www.sanger.ac.uk/Software/Pfam/, in Sweden at http://www.cgb.ki.se/Pfam/, in France at http://pfam.jouy.inra.fr/ and in the US at http://pfam.wustl.edu/. The latest version (6.6) of Pfam contains 3071 families, which match 69% of proteins in SWISS-PROT 39 and TrEMBL 14. Structural data, where available, have been utilised to ensure that Pfam families correspond with structural domains, and to improve domain-based annotation. Predictions of non-domain regions are now also included. In addition to secondary structure, Pfam multiple sequence alignments now contain active site residue mark-up. New search tools, including taxonomy search and domain query, greatly add to the functionality and usability of the Pfam resource.
Figures
References
-
- Sonnhammer E.L.L., Eddy,S.R. and Durbin,R. (1997) Pfam: A comprehensive database of protein domain families based on seed alignments. Proteins, 28, 405–420. - PubMed
-
- Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. - PubMed
-
- Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. - PubMed
-
- Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D. et al. (2000) InterPro—an integrated documentation resource for protein families, domains and functional sites. Bioinformatics, 16, 1145–1150. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

