Femtosecond dynamics of rubredoxin: tryptophan solvation and resonance energy transfer in the protein
- PMID: 11752400
- PMCID: PMC117505
- DOI: 10.1073/pnas.012582399
Femtosecond dynamics of rubredoxin: tryptophan solvation and resonance energy transfer in the protein
Abstract
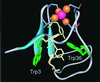
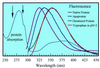
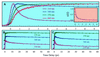
We report here studies of tryptophan (Trp) solvation dynamics in water and in the Pyrococcus furiosus rubredoxin protein, including the native and its apo and denatured forms. We also report results on energy transfer from Trp to the iron-sulfur [Fe-S] cluster. Trp fluorescence decay with the onset of solvation dynamics of the chromophore in water was observed with femtosecond resolution ( approximately 160 fs; 65% component), but the emission extended to the picosecond range (1.1 ps; 35% component). In contrast, the decay is much slower in the native rubredoxin; the Trp fluorescence decay extends to 10 ps and longer, reflecting the local rigidity imposed by residues and by the surface water layer. The dynamics of resonance energy transfer from the two Trps to the [Fe-S] cluster in the protein was observed to follow a temporal behavior characterized by a single exponential (15-20 ps) decay. This unusual observation in a protein indicates that the resonance transfer is to an acceptor of a well-defined orientation and separation. From studies of the mutant protein, we show that the two Trp residues have similar energy-transfer rates. The critical distance for transfer (R(0)) was determined, by using the known x-ray data, to be 19.5 A for Trp-36 and 25.2 A for Trp-3, respectively. The orientation factor (kappa(2)) was deduced to be 0.13 for Trp-36, clearly indicating that molecular orientation of chromophores in the protein cannot be isotropic with kappa(2) value of 2/3. These studies of solvation and energy-transfer dynamics, and of the rotational anisotropy, of the wild-type protein, the (W3Y, I23V, L32I) mutant, and the fmetPfRd variant at various pH values reveal a dynamically rigid protein structure, which is probably related to the hyperthermophilicity of the protein.
Figures



Similar articles
-
A comparative picosecond-resolved fluorescence study of tryptophan residues in iron-sulfur proteins.FEBS Lett. 1994 Jul 18;348(3):305-10. doi: 10.1016/0014-5793(94)00606-7. FEBS Lett. 1994. PMID: 8034060
-
Biological water at the protein surface: dynamical solvation probed directly with femtosecond resolution.Proc Natl Acad Sci U S A. 2002 Feb 19;99(4):1763-8. doi: 10.1073/pnas.042697899. Epub 2002 Feb 12. Proc Natl Acad Sci U S A. 2002. PMID: 11842218 Free PMC article.
-
A neutron crystallographic analysis of a rubredoxin mutant at 1.6 A resolution.Acta Crystallogr D Biol Crystallogr. 2004 Aug;60(Pt 8):1364-73. doi: 10.1107/S090744490401176X. Epub 2004 Jul 21. Acta Crystallogr D Biol Crystallogr. 2004. PMID: 15272158
-
W3Y single mutant of rubredoxin from Pyrococcus furiosus: a preliminary time-of-flight neutron study.Acta Crystallogr D Biol Crystallogr. 2004 Jan;60(Pt 1):200-2. doi: 10.1107/s0907444903024041. Epub 2003 Dec 18. Acta Crystallogr D Biol Crystallogr. 2004. PMID: 14684930
-
Neutron diffraction studies on rubredoxin from Pyrococcus furiosus.J Synchrotron Radiat. 2004 Jan 1;11(Pt 1):76-9. doi: 10.1107/s0909049503024178. Epub 2003 Nov 28. J Synchrotron Radiat. 2004. PMID: 14646139
Cited by
-
Validation of response function construction and probing heterogeneous protein hydration by intrinsic tryptophan.J Phys Chem B. 2012 Nov 15;116(45):13320-30. doi: 10.1021/jp305118n. Epub 2012 Nov 2. J Phys Chem B. 2012. PMID: 23075091 Free PMC article.
-
Dissection of complex protein dynamics in human thioredoxin.Proc Natl Acad Sci U S A. 2007 Mar 27;104(13):5366-71. doi: 10.1073/pnas.0608498104. Epub 2007 Mar 16. Proc Natl Acad Sci U S A. 2007. PMID: 17369362 Free PMC article.
-
Hydration at the surface of the protein Monellin: dynamics with femtosecond resolution.Proc Natl Acad Sci U S A. 2002 Aug 20;99(17):10964-9. doi: 10.1073/pnas.162366099. Epub 2002 Aug 12. Proc Natl Acad Sci U S A. 2002. PMID: 12177425 Free PMC article.
-
Water Determines the Structure and Dynamics of Proteins.Chem Rev. 2016 Jul 13;116(13):7673-97. doi: 10.1021/acs.chemrev.5b00664. Epub 2016 May 17. Chem Rev. 2016. PMID: 27186992 Free PMC article. Review.
-
Protein surface hydration mapped by site-specific mutations.Proc Natl Acad Sci U S A. 2006 Sep 19;103(38):13979-84. doi: 10.1073/pnas.0606235103. Epub 2006 Sep 12. Proc Natl Acad Sci U S A. 2006. PMID: 16968773 Free PMC article.
References
-
- Szabo A G, Rayner D M. J Am Chem Soc. 1980;102:554–563.
-
- Petrich J W, Chang M C, McDonald D B, Fleming G R. J Am Chem Soc. 1983;105:3824–3832.
-
- Beechem J M, Brand L. Annu Rev Biochem. 1985;54:43–71. - PubMed
-
- Ruggiero A J, Todd D C, Fleming G R. J Am Chem Soc. 1990;112:1003–1014.
-
- Callis R R. Methods Enzymol. 1997;278:113–150. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

