The remarkable structural and functional organization of the eukaryotic pyruvate dehydrogenase complexes
- PMID: 11752427
- PMCID: PMC64939
- DOI: 10.1073/pnas.011597698
The remarkable structural and functional organization of the eukaryotic pyruvate dehydrogenase complexes
Abstract
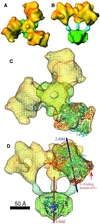
The three-dimensional reconstruction of the bovine kidney pyruvate dehydrogenase complex (M(r) approximately 7.8 x 10(6)) comprising about 22 molecules of pyruvate dehydrogenase (E(1)) and about 6 molecules of dihydrolipoamide dehydrogenase (E(3)) with its binding protein associated with the 60-subunit dihydrolipoamide acetyltransferase (E(2)) core provides considerable insight into the structural and functional organization of the largest multienzyme complex known. The structure shows that potentially 60 centers for acetyl-CoA synthesis are organized in sets of three at each of the 20 vertices of the pentagonal dodecahedral core. These centers consist of three E(1) molecules bound to one E(2) trimer adjacent to an E(3) molecule in each of 12 pentagonal openings. The E(1) components are anchored to the E(1)-binding domain of the E(2) subunits through an approximately 50-A-long linker. Three of these linkers emanate from the outside edges of the triangular base of the E(2) trimer and form a cage around its base that may shelter the lipoyl domains and the E(1) and E(2) active sites. The docking of the atomic structures of E(1) and the E(1) binding and lipoyl domains of E(2) in the electron microscopy map gives a good fit and indicates that the E(1) active site is approximately 95 A above the base of the trimer. We propose that the lipoyl domains and its tether (swinging arm) rotate about the E(1)-binding domain of E(2,) which is centrally located 45-50 A from the E(1), E(2), and E(3) active sites, and that the highly flexible breathing core augments the transfer of intermediates between active sites.
Figures



Similar articles
-
Interaction of avidin with the lipoyl domains in the pyruvate dehydrogenase multienzyme complex: three-dimensional location and similarity to biotinyl domains in carboxylases.Proc Biol Sci. 1992 Jun 22;248(1323):247-53. doi: 10.1098/rspb.1992.0069. Proc Biol Sci. 1992. PMID: 1354363
-
Stoichiometry of binding of mature and truncated forms of the dihydrolipoamide dehydrogenase-binding protein to the dihydrolipoamide acetyltransferase core of the pyruvate dehydrogenase complex from Saccharomyces cerevisiae.Biochemistry. 1996 May 7;35(18):5879-82. doi: 10.1021/bi9600254. Biochemistry. 1996. PMID: 8639549
-
Structure of the native pyruvate dehydrogenase complex reveals the mechanism of substrate insertion.Nat Commun. 2021 Sep 6;12(1):5277. doi: 10.1038/s41467-021-25570-y. Nat Commun. 2021. PMID: 34489474 Free PMC article.
-
Atomic structure of the cubic core of the pyruvate dehydrogenase multienzyme complex.Science. 1992 Mar 20;255(5051):1544-50. doi: 10.1126/science.1549782. Science. 1992. PMID: 1549782 Review.
-
Molecular biology and biochemistry of pyruvate dehydrogenase complexes.FASEB J. 1990 Nov;4(14):3224-33. doi: 10.1096/fasebj.4.14.2227213. FASEB J. 1990. PMID: 2227213 Review.
Cited by
-
Down regulation of the expression of mitochondrial phosphopantetheinyl-proteins in pantothenate kinase-associated neurodegeneration: pathophysiological consequences and therapeutic perspectives.Orphanet J Rare Dis. 2021 May 5;16(1):201. doi: 10.1186/s13023-021-01823-3. Orphanet J Rare Dis. 2021. PMID: 33952316 Free PMC article.
-
Actinobacteria challenge the paradigm: A unique protein architecture for a well-known, central metabolic complex.Proc Natl Acad Sci U S A. 2021 Nov 30;118(48):e2112107118. doi: 10.1073/pnas.2112107118. Proc Natl Acad Sci U S A. 2021. PMID: 34819376 Free PMC article.
-
Gender-Related Effect of Sodium Dichloroacetate on the Number of Hassall's Corpuscles and RNA NKCC1 Expression in Rat Thymus.Biomed Res Int. 2019 Apr 24;2019:1602895. doi: 10.1155/2019/1602895. eCollection 2019. Biomed Res Int. 2019. PMID: 31179315 Free PMC article.
-
Structures of the human pyruvate dehydrogenase complex cores: a highly conserved catalytic center with flexible N-terminal domains.Structure. 2008 Jan;16(1):104-14. doi: 10.1016/j.str.2007.10.024. Structure. 2008. PMID: 18184588 Free PMC article.
-
Markov-State Transition Path Analysis of Electrostatic Channeling.J Phys Chem C Nanomater Interfaces. 2019 Jun 20;123(24):15284-15292. doi: 10.1021/acs.jpcc.9b02844. Epub 2019 May 22. J Phys Chem C Nanomater Interfaces. 2019. PMID: 31275507 Free PMC article.
References
-
- Reed L J, Hackert M L. J Biol Chem. 1990;265:8971–8974. - PubMed
-
- Patel M S, Roche T E. FASEB J. 1990;4:3224–3233. - PubMed
-
- Guest J R, Angier S J, Russell G C. Ann NY Acad Sci. 1989;573:76–99. - PubMed
-
- Perham R N. Annu Rev Biochem. 2000;69:961–1004. - PubMed
-
- Zhou Z H, Liao W, Cheng R H, Lawson J E, McCarthy D B, Reed L J, Stoops J K. J Biol Chem. 2001;276:21704–21713. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

